|
BioDiscovery : Research Poster
|
|
Corresponding author: Lucia Cimarelli (lucia.cimarelli@unicam.it)
Academic editor: Nikolai Zhelev
Received: 10 Oct 2016 | Accepted: 02 Feb 2017 | Published: 08 Feb 2017
© 2017 Lucia Cimarelli, Anna Maria Giuliodori, Anna Brandi, Karolina Adamkiewicz, Roberto Spurio, Attilio Fabbretti
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Cimarelli L, Giuliodori A, Brandi A, Adamkiewicz K, Spurio R, Fabbretti A (2017) Screening an Archetypal Collection of Microorganisms for the Presence of Unexplored Antimicrobial Compounds. BioDiscovery 20: e10763. https://doi.org/10.3897/biodiscovery.20.e10763
|

|
Abstract
Background
WHO estimates that more than 700.000 people die every year as a result of drug-resistant infections. To tackle this problem and change the current trend, it is necessary to design advanced strategies for drug discovery and to promote early-stage research activities finalized at the development of new drugs (
New information
In this study, we present the results of the preliminary screening of a library of microorganisms, collected from different environmental settings. Approximately 300 strains of the culture collection were tested on solid medium for inhibition of growth of three tester species, namely Bacillus subtilis, Escherichia coli and Staphylococcus aureus. The selection of the tester species was made according to the following criteria:
-
Staphylococcus aureus and Escherichia coli were chosen as representative of Gram+ and Gram- bacteria, respectively. Furthermore, these organisms are relevant for the global public health because the number of antibiotic resistant strains responsible for invasive diseases is steadily increasing (
Centers for Disease Control and Prevention 2014 ,Lowy 2003 ); - Bacillus subtilis, instead, was selected as Gram+ tester microorganism commonly found in soil samples. Thus, this well characterized bacterium can be used to gain insight on the effect of metabolites produced by microorganisms of the culture collection described in this study.
One of the active strains, MES18, was classified as a Bacillus spp. by means of small-subunit rRNA gene sequencing. To identify the compound(s) responsible for this inhibitory activity, MES18 cells were grown in liquid medium at 30°C and samples were taken at different time points over a period of 12 days. The supernatants obtained from the fermentation media were subjected to fractionation by chromatography on reversed-phase column and all the eluted compounds were assayed for their ability to repress the growth of tester strains. This approach allowed us to identify the fractions containing the bioactive compound(s) and to establish that the production of these secondary metabolites reached a maximum during the idiophase of the cell culture, when cell growth and replication decline. Further analyses to identify the physical-chemical features of the compound(s) produced by this strain using HPLC coupled to mass-spectrometry are currently ongoing.
Keywords
Drug-resistance, secondary metabolites, antibiotics.
Methods
In this study, more than 900 microorganisms were isolated from different environmental samples. They are maintained in an archetypal culture collection at the University of Camerino (Unicam). A Gram- (Escherichia coli) and two Gram+ microorganisms (Bacillus subtilis and Staphylococcus aureus) were used as tester species for the antibacterial activity investigation on approximately 300 strains of the culture collection. Each microorganism was streaked in a circle at the center of three plates of solid medium. Once the growth was markedly detectable, the three tester species were transferred on these plates by replica plating and the size of the inhibition zone was regularly monitored and measured. Subsequently, to identify the compound(s) responsible for the inhibitory activity, supernatants obtained from the fermentation media were taken at different time points over a period of about two weeks. These samples were subjected to fractionation by reverse-phase chromatography and all the eluted compounds were evaluated by disk-diffusion assay.
Results and discussion
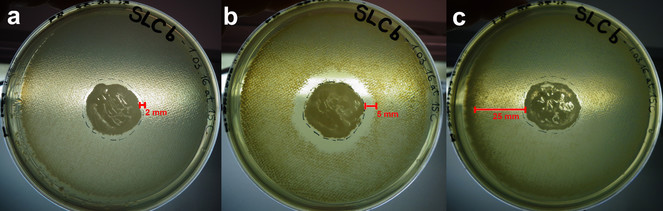
Using this straightforward approach, we identified several microorganisms able to produce secondary metabolites with antimicrobial activity. Here, we present the results of one of the active strains, MES18, isolated from a soil sample next to the Lake of Cingoli in Central Italy. Based on 16S rRNA sequence analysis, MES18 has been placed in the Bacillus spp. group. After five days of incubation, the inhibition displayed by MES18 reached 2mm in the case of Escherichia coli, 5mm in the case of Staphylococcus aureus and a notably reduced growth of Bacillus subtilis, which appears fuzzy over a zone of at least 25mm (Fig.
Halo inhibition assay. LB agar plates with the active microorganism, MES18, tested on three tester species (a E. coli, b S. aureus and c B. subtilis). The inhibition zone is shown by a red line.
Conclusions
In this work, we have presented an investigation carried out on our collection of microorganisms (bacteria, fungi, algae) as part of an ongoing project on the identification of metabolites for bio-industrial use. The effect of metabolites produced by MES18 on three tester species is presented as a proof-of-concept study to highlight the potential repertoire of molecules synthesized by microorganisms. The isolation and characterization of these active molecules is a key step to advance the research on new lead compounds that could be used as template for the development of new drugs. In light of the global challenge of the antibiotic-resistant infections, this research activity should be regarded as a ‘categorical imperative’ (in Kant’s words).
Presented at
II International Conference on Clinical Sciences and Drug Discovery (CSDD 2016, Suppl. material
Funding program
Grant title
New antimicrobial agents targeting antibiotic resistance mechanisms and novel or underexploited targets. RBFR130VS5_001 to Attilio Fabbretti
Hosting institution
University of Camerino, School of Biosciences and Veterinary Medicine, Laboratory of Molecular Biology and Microbial Biotechnology.
References
-
Antibiotic Resistance Threats in the United States, 2013. https://www.cdc.gov/drugresistance/threat-report-2013/
-
Antimicrobial resistance: the example of Staphylococcus aureus.Journal of Clinical Investigation111(9):1265‑1273. https://doi.org/10.1172/jci200318535
Supplementary material
Download file (395.82 kb)