1. Introduction
B-cell chronic lymphocytic leukemia (B-CLL), is the most common leukaemia in the western world. It is not curable [
Although prognostic markers have been identified, there is as yet no proven indications for initiating treatment in patients with asymptomatic disease.. The lack of accuracy in predicting disease progression and survival on an individual basis has been a long asked research question in B-CLL. It’s a known fact that in B-CLL disease pathogenesis to date, no single biomarker has been useful in predicting the prognosis in CLL patients. This review focuses on our current understanding of clinical course and molecular mechanisms in B-CLL disease, and suggests the application of systems biology as an approach in personalized treatment for B-CLL patients.
2. Clinical paradigm
The clinical course of B-CLL disease is highly variable, with life expectancies ranging from months to decades. There is no apparent survival advantage for early treatment intervention, and some patients may never require treatment. At present the information available is only used to counsel patients and to help inform clinicians regarding the frequency of monitoring. The currently available clinical staging systems (e.g. Rai or Binets test) for B-CLL are simple and inexpensive but lack accuracy to predict disease progression and survival on an individual basis. Prediction was not significant when analyzed using life table methods performed by Goldin et al. [
B-CLL disease is characterized by an accumulation of mature, non-proliferating B lymphocytes in the blood, spleen, lymph nodes and bone marrow. The accumulation of B cells is the result of a proliferative defect, failing B cells to undergo apoptosis and leading to large numbers of cells being blocked in the G0/G1 phase of the cell cycle [
The diagnosis of B-CLL is carried out using a number of classic and novel sensitive techniques, which allow diagnosing and differentiating the disease from other chronic lympho proliferative disorders [
B-CLL disease has been presented as two distinct types. The presence of somatic mutations in the immunoglobulin (IgH) heavy chain gene define a group of patients exhibiting stable or slowly progressive disease that requires late or no treatment. By contrast, the absence of mutations in the IgH genes of B-CLL cells define a group, which exhibit a progressive clinical course requiring early treatment [
3. Cell cycle, proliferation and apoptosis
B-CLL lymphocytes lack the fundamental functions of regulating the cell cycle and apoptosis. That is why the cellular factors that govern the entry of resting B-cells into the cell cycle and promote their progression through the different phases of cell cycle, are considered to be optimal targets for drug therapy. Normally the matured naive lymphocytes circulating in blood remain quiescent in the G0 phase and reside in lymphoid tissues such as the spleen until they encounter an appropriate mitogenic stimulus. The progression of the cell cycle in naive B cells depends on the PI3K activation of AKT activation, and inducing NF-kB activation and cyclin D2 activity by deregulating FOXO transcription rate [
The proliferation rate and resistance to drug-induced apoptosis in B cells are recognized as important factors in the outcome of treatment in CLL disease. Mutated and unmutated CLL patients clearly differ in terms of prognosis. The genomic aberrations occurring in B-CLL patients have their individual effects due to distinct expression profiles, as reported by Klein et al [
To date large components of BCR signalling pathways have been elucidated in determining the prognosis of B-CLL, but failed to predict accurately. Pharmacological inhibition of BCR signalling in CLL can be achieved by targeting components of the BCR signalling cascade. Several groups have reported an increase in MAPK (ERK2) phosphorylation and NFAT transcriptional activity and a corresponding lack of AKT phosphorylation due to constitutive activation of distinct BCR signalling pathways in a subset of particular CLL cases [
Increased aggregation of B lymphocytes due to a lack of apoptotic signalling has been a well known cause in B- CLL disease pathogenesis, suggesting that B-CLL cells lack the fine tuning of a balance between pro-apoptotic and anti apoptotic factors in the microenvironment. The anti-apoptotic protein Bcl-2 is highly expressed in B-CLL patients and has been well characterized, but paradoxically, high levels of pro-apoptotic molecules have also been observed [
4. Microenvironment: Cross talk and interplay
Microenvironment regulating signals are crucial for the process of signaling in leukemic cells that induce proliferation and lead to the survival and accumulation of leukemic cells within lymphoid organs. The in vivo accumulation of leukaemic lymphocytes is potentiated by interactions of CLL cells with other cells such as mesenchymal stromal cells (MSCs), nurse-like cells (NLCs), T cells and other soluble factors that include Interleukins (IL-4) and cytokines such as CCL22 and CCL17 [
Common genetic aberrations that occur in CLL disease and different stimuli originating from the microenvironment, cooperate in the selection and expansion of the malignant clone. CLL cells relentlessly accumulate in vivo but rapidly undergo spontaneous apoptosis in vitro. This implies that their apoptosis resistance, rather than being an intrinsic feature of leukemic stage, depends on external signals for survival since the maturation stages of B cells are highly dependent on microenvironment signals [
Exemplified crosstalk between CLL cells and accessory cells occurs in the marrow and/or lymphoid tissue microenvironments. Contact between CLL cells and NLCs or MSCs is established and maintained by chemokine receptors and adhesion molecules. NLCs express the chemokines CXCL12 and CXCL13, whereas MSCs predominantly express CXCL12. NLCs and MSCs attract CLL cells via the G protein coupled chemokine receptors CXCR4 and CXCR5, which are expressed at high levels on CLL cells [
Through this mechanism, CLL cells can actively recruit T cells for cognate T-cell interactions with CLL cells. CD40L T-cells are preferentially found in CLL proliferation centers and can interact with CLL cells via CD40. Cytokines secreted by T-cells or CLL cells such as TNF-α or IL-4 are considered important regulators of CLL cell survival. Moreover, downregulation of proximal and intermediate T-cell receptor signalling cascades and globally reduced cytokine secretion, contributes to significant immunodeficiency in non-leukemic indolent B-cell lymphomas [
5. Micro RNAs
Micro RNA’s (miRNAs/miR’s) are a recently discovered class of molecules that regulate gene expression at the posttranscriptional level. The interactions between miRNAs, target genes, and pathways in CLL are clearly complex, as are the links between genotype and phenotype miRNA machinery, which include miRNAs involved in many cellular processes such as proliferation and apoptosis. Some microRNAs are referred to as oncomiR’s and exhibit differential expression level in cancer disease by acting as either oncogenes or tumour suppressor genes [
Cimmino et al. [
The demonstration of down- regulation of cyclin-E2, CDK6, E2F5, cyclin D1 (CCND1) and Bcl-2 protein levels after induction of miR-34a expression has been noted to be important, suggesting that miR-34a in CLL mediates its functions within and potentially outside the p53 pathway [
Analysis of miRNA expression values between the predefined subgroups showed that the miR-223, the miR-29b, the miR-29c, and the miR-181 family are down regulated in 17p-aggressive cases showing the highest level of Tcl-1 expression compared with 17p-indolent cases [
6. Drug targets and resistance
Adequate progress has been made in recent times for therapeutic intervention and management B-CLL disease. New insights into the molecular pathology of B-CLL have generated a plethora of biological markers that predict the prognosis and influence therapeutic decisions. These markers include historical Rai and Binet staging systems, loss of p53 and ATM functions, the unmutated IgVH gene, and high expression of ZAP-70 or CD38 [
A recent review by García-Escobar et al. [
Additional monoclonal antibodies targeting CD20, CD23, CD37, CD38 or CD40, as well as drugs designed to interfere with proteins regulating the cell cycle, apoptotic machinery or leukemic microenvironment (e.g., flavopiridol, oblimersen, ABT-263 or lenalidomide) are being investigated in clinical trials. To date, single-agent clinical trials have indicated that the major clinical outcome is the stabilization of disease states. Lyse [
The main focus of therapeutic strategies in B-CLL is still cytotoxic chemotherapeutically available alkylating agents or purine analogues that trigger DNA damage response via p53 leading to a prominent cell death. Most of B-CLL diseased patients carry defects in the p53 pathway and therefore it has been challenging to overcome the resistance through p53 independent cell death pathways [
7. Systems biology approach
Although clinical stages such as Rai and Binet tests are the required basis for the prognosis in the B-CLL patients, many other biological markers mentioned in the above section have been offering important insights to prognostic information. Nonetheless these prognostic factors have not been fully validated or standardized in large clinical trials. Moreover, it is practically impossible to validate all possible biomarkers in clinical trials due to the fact that response upon drug dosage in B-CLL patients is varied, and therefore could not sustain in clinical evaluation. Existence of conflicting data in research studies has added more complexity and confusion in the evaluation of B-CLL disease pathogenesis. For example Klein et al [
Notably, Klein et al [
The above mentioned molecular signatures may have a profound impact on prognosis for B-CLL disease pathogenesis, but the lack of routine clinical methods for diagnosis is a major challenge which should be accounted for in future research. Taking into consideration our current understanding of B-CLL disease pathogenesis, it is now convincing that application of systems biology should be employed as an approach for predicting patient response and designing individualized therapies. An iterative interplay between biological experiments producing quantitative data followed by suitable modeling strategies should enable us to interpret the biology in a global system perspective, thereby increasing our current understanding of the signaling pathways involved in the pathogenesis of a complex disease such as B-CLL. This approach should also include the extracellular aspects of the disease mechanism such as microenvironment. Due to the fact of these differential study conclusions, it is now time to consider prognosis and treatment on an individual basis rather than cohort studies involving large numbers of patients. Moreover it is now evident that B-CLL is not a single target regulated disease, but rather has multiple targets with two distinct disease phenomenae (indolent and aggressive forms).
Building network models by applying systems biology is an approach that will enhance our understanding of key molecular players in the B-CLL disease pathogenesis. To generate a computational model for prognosis of B-CLL patients in silico, one has to overcome a number of challenges such as working with a large number of chemically diverse molecules that are often rapidly changing their concentrations in terms of their expression and regulation [
Disease progression can vary considerably from one patient to another suggesting that future therapies will need to be designed on an individual basis. To do this, it requires an in-depth analysis of two major groups of variables. Firstly by encompassing the molecular interactions between the various intra-cellular proteins which implement the cell’s regulatory pathways, in particular survival, anti-growth, proliferation and apoptosis (Figure 1). Secondly, as these interactions are highly complex at the intra-cellular level where the loss of homeostasis has to be analysed, a model reporting on the molecular activity, and the imbalance between B-cell proliferation and apoptosis should be addressed. Due to this complexity, some form of holistic approach to the problem is required and we believe that one way forward is to describe the dynamics of the various molecular and cellular interactions by using a mathematical model. In this way the mechanistic links, which underpin the B-CLL disease phenotype can be defined and, subject to calibration at the individual patient level, these models would enable a better understanding of disease progression, as well as providing a methodology for targeted drug interventions.
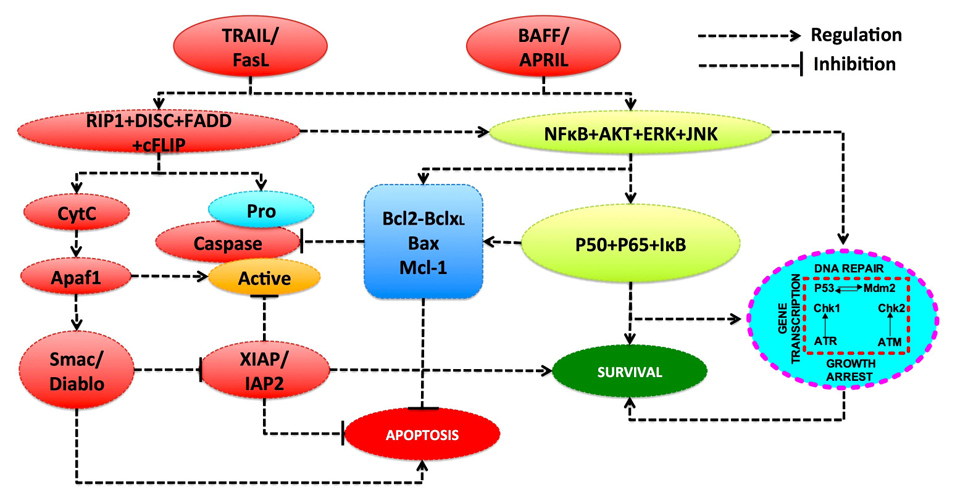
The key aspects in cellular signaling that involve the cell cycle, apoptosis and various intracellular pathways such as DNA damage repair and cytokine receptor activated network activating PI3K/AKT, are considered to be crucial components that are frequently deregulated in B-CLL disease. Each pathway is subject to an intensive study by the computational systems biology approach: modelling of the cell cycle [
The mathematical approach to modelling the cell cycle was developed in a series of works by Novak and Tyson [
7.1 Identification of fragile nodes
In this context a new area in systems biology is being developed that aims at not only the description of the normal functioning of cellular control systems, but also the modelling of the mechanics of their malfunction in pathological conditions including cancer [
One of the powerful methods for determining fragile nodes within cellular signaling networks is sensitivity analysis of the computational models [
The results of sensitivity analysis of the cell cycle models obtained by Nayak S et al. [
![Figure 2. Sensitivity analysis of cell cycle models. Sensitivity coefficients [82] determined for the G1/S model [66] (A) and the G2-DNA damage model [67] (B). Abbreviations: CycE - cyclin E, aCycE/iCycE – active/inactive CycE, CDC25 - a dual-specificity phosphatase CDC25A, aCDC25/iCDC25 – active/inactive CDC25, Cdk2 - cyclin dependent kinase 2, E2F – transcription factor E2F, CKI - a cyclin dependent kinase inhibitor, aCKI/iCKI – active/inactive CKI, pRB - retinoblastoma protein, CycD - cyclin D, Cdk4/6 – cyclin dependent kinase 4 or 6, pMPF - pre-maturation promoting factor, p21 - cyclin-dependent kinase inhibitor, 14-3-3s - 14-3-3s protein, Wee1 - Wee1 kinase. Data are represented by courtesy permission of Prof. J.D. Varner. Figure 2](https://biodiscovery.pensoft.net/showimg/oo_85773.jpg)
The second ranked fragile mechanism in the G1/S network obtained in sensitivity analysis is cyclin E-CDK2 complex. Inhibition of the active cyclin E-CDK2 is being considered as a treatment strategy in different types of cancer including B-CLL. For example, a synthetic flavone, flavopiridol induces cell cycle arrest by inhibiting multiple CDKs, and leads to p53-independent apoptosis in CLL cells [
7.2 Signalling simulation models
The signaling pathway relevant to B-CLL disease is PI3K/AKT pathway, which plays a key role in cell proliferation, differentiation and survival [
Sensitivity analysis of these models being a powerful method for analysing PI3K/AKT signaling in cancer was carried out in many works to identify fragile points in this pathway [
The results of sensitivity analysis of the PI3K/AKT pathway model [
![Figure 3. Sensitivity analysis of the PI3K/AKT signaling model [91]. Representation of sensitivity of phosho-AKT signal to kinetic parameters: rate constant ki, dissociation constants Kd,i, maximal rate of reaction Vm,i, and Michaelis-Menten constants, Km,i (A) and to the initial concentrations of the enzymes (B). Abbreviations: AKT - protein kinase B, pAKT – phosphorylated AKT, PI3K - phosphatidylinositol-3-kinase, PTEN - phosphatase and tensin homolog, PDK1 - phosphoinositide-dependent protein kinase-1, PP2A - protein phosphatase 2A, PIP3 - phosphatidylinositol (3,4,5)-trisphosphate, PIP2 - phosphatidylinositol 4,5-bisphosphate. Figure 3](https://biodiscovery.pensoft.net/showimg/oo_85774.jpg)
7.3 Apoptosis simulation models
The high sensitivity of PI3K/AKT signalling to AKT expression level revealed in sensitivity analysis (see figure 3B) showed its significant impact to anti-apoptotic signal. Activated AKT inhibits apoptosis by phosphorylation of Bad protein, procaspase-9, and involves in NF-kB activation, so AKT inhibition is an attractive target for CLL drug therapy. Preclinical trials showed that AKT inhibitor, A-443654 induces apoptosis in B-CLL cells [
A large number of theoretical studies are devoted to computational modelling of both the intrinsic and extrinsic apoptotic pathways [
The extrinsic apoptosis is triggered by binding the tumour necrosis factor receptors (TNF-Rs) with members of TNF family ligands (TNF-α, FasL, and TRAIL) [
The high level of sensitivity of the model output (caspase-3 activation) showed the following sensitive points of this pathway: 1. kinetic parameters of TRAIL ligand-TRAIL receptor interaction and concentrations of membrane receptors; 2. activation of pro-apoptotic protein Bak/Bax negatively regulated by Bcl-2 protein; and 3. Smac/XIAP inhibition of caspase-3 and caspase-9 resulting in positive feedback in apoptosis. The sensitivity of caspase-3 activation to Bcl-2 concentration was obtained at upregulation of Bcl-2 as opposed to insensitivity to Bcl-2 at downregulation of Bcl-2 when Bcl-2 does not inhibit the type II apoptotic pathway significantly (see the representation of the sensitivity coefficients at the decrease and increase of protein expression levels in figure 4B) [
High sensitivity of caspase-3 activation with the affinity of TRAIL ligand to TRAIL receptor and their concentrations obtained in sensitivity analysis (figure 4), correlates with a significant role of the death receptor in initiation of apoptosis in B-CLL cells [
![Figure 4. Sensitivity analysis of the apoptosis models. (A) Results of sensitivity analysis of the model of TRAIL-induced apoptosis to kinetic parameters of the model: the rates of the forward, kf and reverse reactions, kr [108]. (B) Results of sensitivity analysis of the model of FasL-induced apoptosis to concentration of the proteins involved [69]. Data represent the change in the half-time of caspase-3 activation (hour) at the variation of protein level two orders of magnitude in either direction from the baseline values. Abbreviations: Fas - FAS receptor, FasL- Fas ligand; XIAP - X-linked inhibitor of apoptosis protein, FADD - Fas-associated protein with death domain, casp3 - a caspase-3 protein, casp9 - a caspase-9 protein, casp8 - a caspase-8 protein, cytoC - cytochrome c, FLIP - flice inhibitory protein, Bax - Bcl-2 associated X protein, Bcl-2 - B-cell lymphoma 2 protein, Bid - BH3 interacting domain death agonist, TRAIL- TNF-related apoptosis-inducing ligand, TRAIL-R – TRAIL receptor, Smac - second mitochrondria-derived activator of caspase, Apaf-1 - apoptotic protease activating factor-1. Data are represented by kind permission of Prof. Z. Sun (A) and Prof. D.A. Lauffenburger (B). Figure 4](https://biodiscovery.pensoft.net/showimg/oo_85775.jpg)
The decision making in cell survival/apoptosis is determined by the balance between these competing signals. Two ligands that belong to the TNF superfamily, BAFF (B-cell–activating factor), APRIL (a proliferation-inducing ligand), and their receptors, were reported to be key regulators of B-CLL cell survival [
8. Concluding remarks and perspectives
Building regulatory network model is based on establishing the dynamics of interaction networks showing regulatory pathways involved in gene regulation during the cell cycle and apoptosis. These network models only form a very small part in terms of calibrating of the models, this can be done using patient samples to produce data in terms of the quantitative measurement of key regulatory proteins and cytokine profiling. Once calibrated on an individual patient basis, the models would then enable monitoring of that patient to determine significant changes in the molecular phenotype and, in terms of therapy, permit intervention therapies to be designed using a broad range of bio-therapeutic drugs. A multi-target approach would be possible by identifying key regulators that over come drug resistance trigger apoptosis. Moreover by determining sensitivity nodes or fragile points to induce apoptosis in B-CLL cells, can be useful as a robust drug sensitivity reporter model. These models can result in achieving maximum benefit in treatment regimes for B-CLL patients. Due to the inherent complexity however, particularly in regard to the number of regulatory feedback loops, we believe that a successful approach can only be achieved using mathematical methods.
Modelling the cell cycle has been shown to be extremely robust in terms of the parameter variations. Experimental and clinical data support the hypothesis that computationally identified fragile interactions in the cell cycle represent promising targets for drug therapy [
Acknowledgments
The authors would like to acknowledge financial support from the Northwood Charitable Trust and the Scottish Informatics and Computer Science Alliance (SICSA).
References
- Rozman C, Montserrat E. Chronic lymphocytic leukemia, New Engl J Med 1995; 333: 1052–1057.
Reference Link - Bergsagel DE. The chronic leukemias: a review of disease manifestations and the aims of therapy. Can Med Assoc J 1967; 96: 1615-1620.
- Gunz FW, Fitzgerald PH, Adams A. An abnormal chromosome in chroninc lymphocytic leukaemia. Brit Med J 1962; 2: 1097-1099.
Reference Link - Dameshek W, Savitz HA, Arbor B. Chronic lymphocytic leukemia in twin brothers aged fifty-six. JAMA 1929; 92: 1348-1349.
Reference Link - Branda RF, Ackerman SK, Handwerger BS, Howe RB, Douglas SD. Lymphocyte studies in familial chronic lymphatic leukemia. Am J Med 1978; 64: 508-514.
Reference Link - Furbetta D, Solinas P. Hereditary chronic lymphatic leukemia. Proc 2nd Int Cong Hum Genet. 1963; 2:1078-1079.
- Conley CL, Misiti J, Laster AJ. Genetic factors predisposing to chronic lymphocytic leukemia and to autoimmune disease. Medicine 1980; 59:323-334.
Reference Link - Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood 2004; 104: 1850-1854.
Reference Link - Anaissie EJ, Kontoyiannis DP, O’Brien S, Kantarjian H, Robertson L, Lerner S et al. Infections in patients with chronic lymphocytic leukemia treated with fludarabine. Ann Intern Med 1998; 129: 559-566.
- Hensel M, Kornacker M, Yammeni S, Egerer G, Ho AD. Disease activity and pretreatment, rather than hypogammaglobuli- naemia, are major risk factors for infectious complications in patients with chronic lymphocytic leukaemia. Brit J Haematol 2003; 122: 600-606.
Reference Link - Perkins JG, Flynn JM, Howard RS, Byrd JC. Frequency and type of serious infections in fludarabine-refractory B-cell chronic lymphocytic leukemia and small lymphocytic lymphoma: implications for clinical trials in this patient population. Cancer 2002; 94: 2033-2039.
Reference Link - Francis S, Karanth M, Pratt G, Starczynski J, Hooper L, Fegan C et al. The effect of immunoglobulin VH gene mutation status and other prognostic factors on the incidence of major infections in patientswith chronic lymphocytic leukemia. Cancer 2006; 107: 1023–1033.
Reference Link - Rossi D, De Paoli L, Rossi FM, Cerri M, Deambrogi C, Rasi S et al. Early stage chronic lymphocytic leukaemia carrying unmutated IGHV genes is at risk of recurrent infections during watch and wait. Br J Haematol 2008; 141: 734-736.
Reference Link - Hamblin TJ, Oscier DG. Chronic lymphocytic leukaemia: the nature of the leukaemic cell. Blood Rev 1997; 11: 119-128.
Reference Link - MacFarlane M, Harper N, Snowden TR, Dyer MJ, Barnett GA, Pringle JH et al. Mechanisms of resistance to TRAIL-induced apoptosis in primary B cell chronic lymphocytic leukaemia. Oncogene 2002; 21: 6809-6818.
Reference Link - Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O'Brien S et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood 1996; 87(12): 4990-4997.
- Todorov IT, Philipova RN, Zhelev NZ, Hadjiolov AA. Monoclonal antibody to a nucleolar antigen of human B-lymphoblastoid cells. Cell Biol Int Rep 1987; 11(3): 181-187.
Reference Link - Matutes E, Attygalle A, Wotherspoon A, Catovsky D. Diagnostic issues in chronic lymphocytic leukaemia (CLL). Best Pract Res Clin Haematol 2010; 23(1): 3-20.
Reference Link - Staudt LM. Molecular diagnosis of the hematologic cancers. New Eng J Med 2003; 348: 1777-1785.
Reference Link - Grumont RJ, Strasser A, Gerondakis S. B cell growth is controlled by phosphatidylinosotol 3-kinase dependent induction of Rel/NF-kappaB regulated c-myc transcription. Mol Cell 2002; 10(6): 1283-1294.
Reference Link - Yusuf I, Zhu X, Kharas MG, Chen J, Fruman DA. Optimal B-cell proliferation requires phosphoinositide 3-kinase-dependent inactivation of FOXO transcription factors. Blood 2004; 104(3): 784-787.
Reference Link - Klein U, Dalla-Favera R. New insights into the phenotype and cell derivation of B cell chronic lymphocytic leukemia. Curr Top Microbiol Immunol 2005; 294 31-49.
Reference Link - Haslinger C, Schweifer N, Stilgenbauer S, Döhner H, Lichter P, Kraut N et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol 2004; 22: 3937-3949.
- Gottardi D, Alfarano A, De Leo AM, Stacchini A, Aragno M, Rigo A. et al. In leukaemic CD5 þ B cells the expression of BCL-2 gene family is shifted toward protection from apoptosis. Br J Haematol 1996; 94: 612-618.
Reference Link - Kitada S, Andersen J, Akar S, Zapata JM, Takayama S, Krajewski S et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia correlations with in vitro and in vivo chemoresponses. Blood 1998; 91: 3379-3389.
- Pepper C, Lin TT, Pratt G, Hewamana S, Brennan P, Hiller L et al. Mcl-1 expression has in vitro and in vivo significance in chronic lymphocytic leukemia and is associated with other poor prognostic markers. Blood 2008; 112: 3807-3817.
Reference Link - Veronese L, Tournilhac O, Verrelle P, Davi F, Dighiero G, Chautard E et al. Low MCL-1 mRNA expression correlates with prolonged survival in B-cell chronic lymphocytic leukemia. Leukemia 2008; 22:1291-1293.
Reference Link - Chen R, Plunkett W. Strategy to induce apoptosis and circumvent resistance in chronic lymphocytic leukaemia. Best Pract Res Clin Haematol 2010; 23: 155-166.
Reference Link - Muzio M, Apollonio B, Scielzo C, Frenquelli M, Vandoni I, Boussiotis V et al. Constitutive activation of distinct BCR-signaling pathways in a subset of CLL patients: a molecular signature of energy. Blood 2008; 112:188-195.
Reference Link - Moore VDG, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest 2007; 117: 112-121.
Reference Link - Del GM, Letai A. Rational design of therapeutics targeting the BCL-2 family: are some cancer cells primed for death but waiting for a final push? Adv Exp Med Biol 2008; 615: 159-175.
Reference Link - Kalla C, Scheuermann MO, Kube I, Schlotter M, Mertens D, Döhner H et al. Analysis of 11q22-q23 deletion target genes in B-cell chronic lymphocytic leukaemia: evidence for a pathogenic role of NPAT, CUL5, and PPP2R1B. Eur J Cancer 2007; 43: 1328-1335.
- Tumilasci VF, Oliere S, Nguyen TL, Shamy A, Bell J, Hiscott J. Targeting the apoptotic pathway with BCL-2 inhibitors sensitizes primary chronic lymphocytic leukemia cells to vesicular stomatitis virus induced oncolysis. J Virol 2008; 82(17): 8487-8499.
Reference Link - Zhong F, Harr MW, Bultynck G, Monaco G, Parys JB, De Smedt H et al. Induction of Ca(2)+-driven apoptosis in chronic lymphocytic leukemia cells by peptide-mediated disruption of Bcl-2-IP3 receptor interaction. Blood 2011; 117: 2924-2934.
Reference Link - Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood 2009; 114: 3367-3375.
Reference Link - Ghia P, Chiorazzi N, Stamatopoulos K. Microenvironmental influences in chronic lymphocytic leukaemia: the role of antigen stimulation. J Intern Med 2008; 264: 549-562.
Reference Link - Christopoulos P, Pfeifer D, Bartholome K, Follo M, Timmer J, Fisch P et al. Definition and characterization of the systemic T-cell dysregulation in untreated indolent B-cell lymphoma and very early CLL. Blood 2011; 117: 3836-3846.
Reference Link - Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259-269.
Reference Link - Slack FJ, Weidhaas JB. MicroRNAs as a potential magic bullet in cancer. Future Oncol 2006; 2: 73-82.
Reference Link - Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 2007; 109: 4944-4951.
Reference Link - Visone R, Croce CM. MiRNAs and cancer. Am J Pathol 2009; 174: 1131-1138.
Reference Link - Ward BP, Tsongalis GJ, Kaur P. MicroRNAs in chronic lymphocytic leukemia. Exp Mol Pathol 2011; 90: 173-178.
Reference Link - Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005; 102: 13944-13949.
Reference Link - Zenz T, Mohr J, Eldering E, Kater AP, Bühler A, Kienle D et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 2009; 113: 3801-3808.
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE et al. p53-mediated activation of miRNA34 candidate tumor suppressor genes. Curr Biol 2007; 17: 1298-1307.
Reference Link - Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 2008; 582:1564-1568.
Reference Link - Asslaber D, Pinon JD, Seyfried I, Desch P, Stocher M, Tinhofer I et al. MicroRNA-34aexpression correlates with MDM2 SNP309 polymorphism and treatment-free survival in chronic lymphocytic leukemia. Blood 2010; 115 4191-4197.
Reference Link - Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res 2006; 66 11590- 11593.
Reference Link - Ibrahim S, Keating M, Do KA, O'Brien S, Huh YO, Jilani I et al. CD38 expression as an important prognostic factor in B-cell chronic lymphocytic leukemia. Blood 2001; 98: 181–186.
Reference Link - Węsierska-Gądek J, Krame MP, The impact of CDK inhibition in human malignancies associated with pronounced defects in apoptosis: advantages of multi-targeting small molecules. Future Med Chem 2012; 4(4): 395-424.
Reference Link - McClue SJ, Blake D, Clarke R, Cowan A, Cummings L, Fischer PM et al. In vitro and in vivo antitumor properties of the cyclin dependent kinase inhibitor CYC202 (R-roscovitine). Int J Cancer 2002; 102(5): 463-468.
Reference Link - Whittaker S, Walton M, Kelland L, Garrett M, Zhelev N, Workman P. RB phosphorylation as a pharmacodynamic marker of roscovitine (CYC202) activity in vitro and in vivo. P Am Assoc Canc Res 2001; 42: 926.
- McClue S, Fischer PM, Blake D, Clarke R, Duff S, Krauss E et al. Studies on the mechanism of action of CYC202 (R-roscovitine). P Am Assoc Canc Res 2002; 43: 666.
- García-Escobar I, Sepúlveda J, Castellano D, Cortés-Funes H. Therapeutic management of chronic lymphocytic leukaemia State of the art and future perspectives. Crit Rev Oncol Hematol 2011; 80: 100-113.
Reference Link - Robak T. Recent progress in the management of chronic lymphocytic leukemia, Cancer Treat Rev 2007; 33: 710–728.
Reference Link - Ricci F, Tedeschi A, Morra E, Montillo M. Fludarabine in the treatment of chronic lymphocytic leukemia: a review. Ther Clin Risk Manag 2009; 5: 187-207.
- Norian LA, Kucaba TA, Earel JK, Knutson T, Vanoosten RL, Griffith TS. Synergistic Induction of Apoptosis in Primary B-CLL Cells after Treatment withRecombinant Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand and Histone Deacetylase Inhibitors. J Oncol 2009; 2009: 408038.
- Niedermeier M, Hennessy BT, Knight ZA, Henneberg M, Hu J, Kurtova AV et al. Isoform- selective phosphoinositide3'-kinase inhibitors inhibit CXCR4 signaling and overcome stromal cell-mediated drug resistance in chronic lymphocytic leukemia: a novel therapeutic approach. Blood 2009; 113: 5549-5557.
Reference Link - Herman SE, Gordon AL, Wagner AJ, Heerema NA, Zhao W, Flynn JM et al. The phosphatidylinositol 3-kinase-delta inhibitor CAL-101 demonstrates promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood 2010; 116: 2078-2088.
Reference Link - Pleyer L, Egle A, Hartmann TN, Greil R. Molecular and cellular mechanisms of CLL: novel therapeutic approaches. Nat Rev Clin Oncol 2009; 6: 405-418.
Reference Link - Hartmann TN, Pleyer L, Desch P, Egle A, Greil R. Novel therapeutics approaches to chronic lymphocytic leukemia based on recent biological insights. Discov Med 2009; 8: 157-164.
- Stankovic T, Weber P, Stewart G, Bedenham T, Murray J, Byrd PJ. et al. Inactivation of ataxia telangiectasia mutated gene in B-cell chronic lymphocytic leukaemia. Lancet 1999; 353: 26–29.
Reference Link - Schaffner C, Idler I, Stilgenbauer S, Dohner H, Lichter P. Mantle cell lymphoma is characterized by inactivation of the ATM gene. Proc Natl Acad Sci USA 2000; 97: 2773–2778.
Reference Link - Stankovic T, Hubank M, Cronin D, Stewart GS, Fletcher D, Bignell CR et al. Microarray analysis reveals that TP53- and ATM- mutant B-CLLs share a defect in activating pro- apoptotic responses after DNA damage but are distinguished by major differences in activating prosurvival responses. Blood 2004; 103: 291-300.
Reference Link - Kim YC, Jung YC, Chen J, Alhasan AH, Kaewsaard P, Zhang Y. Evidences showing wide presence of small genomic aberrations in chronic lymphocytic leukemia. BMC Res Notes 2010; 3: 341.
Reference Link - Faratian D, Clyde RG, Crawford JW, Harrison DJ. Systems pathology - taking molecular pathology into a new dimension. Nat Rev Clin Oncol 2009; 6: 455-464.
Reference Link - Dohner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. New Eng J Med 2000; 343: 1910-1916.
- Schrattenholz A, Groebe K, Soskic V. Systems biology approaches and tools for analysis of interactomes and multi-target drugs. Methods Mol Biol 2010; 662: 29-58.
Reference Link - Zhang GJ, Safran M, Wei W, Sorensen E, Lassota P, Zhelev N et al. Bioluminescent imaging of Cdk2 inhibition in vivo. Nat Med 2004; 10: 643-648.
Reference Link - Novak M, Tyson JJ. A model for restriction point control of the mammalian cell cycle. J Theor Biol 2004; 230: 563–579.
Reference Link - Qu Z, Weiss JN, MacLellan WR. Regulation of the mammalian cell cycle: a model of the G1-to-S transition. Am J Physiol-Cell Physiol 2003; 284: C349-C364
- Aguda BD. A qualitative analysis of the kinetics of the G2 DNA damage checkpoint system. Proc Natl Acad Sci USA 1999; 96: 11352–11357.
Reference Link - Clyde RG, Tummala H, Khalil HS, Goszcz K, Lucka I, Tupone MG et al. A novel quantitative systems biology approach to cancer research and treatment. Curr Opin Biotech 2011; 22(S1): S58.
Reference Link - Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell 2011; 144: 926-939.
Reference Link - Hua F, Cornejo MG, Cardone MH, Stokes CL, Lauffenburger DA. Effects of Bcl-2 Levels on Fas Signaling-Induced Caspase-3 Activation: Molecular Genetic Tests of Computational Model Predictions. J Immunol 2005; 175: 985-995.
- Shoemaker JE, Doyle III FJ. Identifying Fragilities in Biochemical Networks: Robust Performance Analysis of Fas Signaling-Induced Apoptosis. Biophys J 2008; 95: 2610–2623.
Reference Link - Birtwistle MR, Hatakeyama M, Yumoto N, Ogunnaike BA, Hoek JB, Kholodenko BN. Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol Syst Biol 2007; 3: 144.
Reference Link - Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie L et al. Therapeutically Targeting ErbB3: A Key Node in Ligand-Induced Activation of the ErbB Receptor–PI3K Axis. Sci Signal 2009; 2(77):) ra31.
- Schoeberl B, Faber AC, Li D, Liang MC, Crosby K, M. Onsum et al. An ErbB3 antibody, MM-121, is active in cancers with ligand-dependent activation. Cancer Res 2010; 70: 2485-2494.
Reference Link - Kitano H. Cancer as a robust system: implications for anticancer therapy. Nat Rev Cancer 2004; 4: 227-235.
Reference Link - Kitano H. A robustness based approach to systems-oriented drug design. Nat Rev Drug Discovery 2007; 6: 202-210.
Reference Link - Nayak S, Salim S, Luan D, Zai M, Varner JD. A test of highly optimized tolerance reveals fragile cell-cycle mechanisms are molecular targets in clinical cancer trials. PLoS One 2008; 3: e2016.
Reference Link - Novak B, Tyson J. Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J Cell Sci 1993; 106: 1153-1168.
- Tyson J, Chen K, Novak B. Network dynamics and cell physiology. Nat Rev Mol Cell Biol 2001; 2: 908-916.
Reference Link - Clyde RG, Bown JL, Hupp TR, Zhelev N, Crawford JW. The role of modelling in identifying drug targets for diseases of the cell cycle. J R Soc Interface 2006; 3: 617-627.
Reference Link - Shiraishi T, Matsuyama S, Kitano H. Large-scale analysis of network bistability for human cancers. PLoS Comput Biol 2010; 6: e1000851.
Reference Link - Badoux XC, Keating MJ, Wierda WG. What is the best frontline therapy for patients with CLL and 17p deletion? Curr Hematol Malig Rep 2011; 6: 36-46.
Reference Link - Varterasian ML, Mohammad RM, Eilender DS, Hulburd K, Rodriguez DH, P.A. Pemberton PA et al. Phase I study of bryostatin 1 in patients with relapsed non-Hodgkin’s lymphoma and chronic lymphocytic leukemia. J Clin Oncol 1998; 16: 56–62.
- Wierda WG, Chiorazzi N, Dearden C, Brown JR, Montserrat E, Shpall E et al. Chronic lymphocytic leukemia: new concepts for future therapy. Clin Lymphoma Myeloma Leuk 2010; 10:369-378.
Reference Link - Kholodenko BN, Demin OV, Moehren G, Hoek JB. Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem 1999; 274: 30169-30181.
Reference Link - Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA et al. Input–output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol 2009; 5 239.
Reference Link - Wang CC, Cirit M, Haugh JM. PI3K-dependent cross-talk interactions converge with Ras as quantifiable inputs integrated by Erk. Mol Syst Biol 2009; 5 246.
Reference Link - Kreeger PK, Lauffenburger DA. Cancer systems biology: a network modeling perspective. Carcinogenesis 2010; 31: 2-8.
Reference Link - Orton RJ, Adriaens ME, Gormand A, Sturm OE, Kolch W, Gilbert DR, Computational modelling of cancerous mutations in the EGFR/ERK signalling pathway. BMC Syst Biol 2009; 3 100.
Reference Link - Faratian D, Goltsov A, Lebedeva G, Moodie S, Mullen P, Kay C et al. Systems biology reveals new strategies for personalising cancer medicine and confirms PTENs role in resistance to trastuzumab. Cancer Res 2009; 69(16): 6713-6720.
Reference Link - Lebedeva G, Sorokin A, Faratian D, Mullen P, Goltsov A, Langdon SP et al. Model-based global sensitivity analysis as applied to identification of anti-cancer drug targets and biomarkers of drug resistance in the ErbB2/3 network. Eur J Pharm Sci 2011; 46(4): 244-258.
Reference Link - Goltsov A, Faratian D, Langdon SP, Harrison DJ, Bown J. Features of the reversible sensitivity-resistance transition in ERK/PI3K/PTEN/AKT signalling network at HER2 inhibition. Cell Signal 2012; 24: 493-504.
Reference Link - Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme, Oncogene 2008; 27: 5497-5510.
Reference Link - Martelli AM, Evangelisti C, Chiarini F, Grimaldi C, Cappellini A, Ognibene A et al. The emerging role of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin signaling network in normal myelopoiesis and leukemogenesis. Biochim Biophys Acta 2010; 1803: 991-1002.
- Niedermeier M, Hennessy BT, Knight ZA, Henneberg M, Hu J, Kurtova AV et al. Burger, Isoform-selective hosphoinositide 3'-kinase inhibitors inhibit CXCR4 signaling and overcome stromal cellmediated drug resistance in chronic lymphocytic leukemia: a novel therapeutic approach. Blood 2009; 113: 5549-5557.
Reference Link - Masood A, Sher T, Paulus A, Miller KC, Chitta S, Chanan-Khan A. Targeted treatment for chronic lymphocytic leukemia. Onco Targets Ther 2011; 4: 169-183.
- De Frias M, Iglesias-Serret D, Cosialls AM, Coll-Mulet L, Santidrián AF, González-Gironès DM et al. Akt inhibitors induce apoptosis in chronic lymphocytic leukemia cells. Haematologica 2009; 94: 1698-1707.
- Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H, Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J Cell Biol 2004; 166: 839-851.
Reference Link - Toettcher JE, Loewer A, Ostheimer GJ, Yaffe MB, Tidor B, Lahav G. Distinct mechanisms act in concert to mediate cell cycle arrest. Proc Natl Acad Sci USA 2009; 106: 785-790.
Reference Link - Aldridge BB, Gaudet S, Lauffenburger DA, Sorger PK. Lyapunov exponents and phase diagrams reveal multi-factorial control over TRAIL-induced apoptosis, Mol Syst Biol 2011; 7: 553.
Reference Link - Komorowski M, Costa MJ, Rand DA, Stumpf MP. Sensitivity, robustness, and identifiability in stochastic chemical kinetics models. Proc Natl Acad Sci USA 2011; 108: 8645-8650.
Reference Link - Koh G, Lee DY. Mathematical modeling and sensitivity analysis of the integrated TNFα-mediated apoptotic pathway for identifying key regulators. Comput Biol Med 2011; 41: 512-528.
Reference Link - Sun T, Yang W, Liu J, Shen P. Modeling the basal dynamics of p53 system. PLoS One 2011; 6: e27882.
Reference Link - Harrington HA, Ho KL, Ghosh S, Tung KC. Construction and analysis of a modular model of caspase activation in apoptosis. Theor Biol Med Model 2008; 5: 26.
Reference Link - F Hua , S. Hautaniemi, R. Yokoo, D.A. Lauffenburger, Integrated mechanistic and data-driven modelling for multivariate analysis of signalling pathways. J R Soc Interface 2006; 3: 515-526.
Reference Link - Perumal TM, Gunawan R. Understanding dynamics using sensitivity analysis: caveat and solution. BMC Syst Biol 2011; 5 41.
Reference Link - Legewie S, Bluthgen N, Herzel H. Mathematical modeling identifies inhibitors of apoptosis as mediators of positive feedback and bistability. PLoS Comput Biol 2006; 2: e120.
Reference Link - Cho KH, Shin SY, Kolch W, Wolkenhauer O. Experimental design in systems biology, based on parameter sensitivity analysis using Maonte Carlo method: a case study for the TNF-mediated NF-B signal transduction pathway. Simulation 2003; 79: 726.
Reference Link - Zhang T, Wu M, Chen Q, Sun Z. Investigation into the regulation mechanisms of TRAIL apoptosis pathway by mathematical modelling. Acta Biochim. Biophys Sin (Shanghai) 2010; 42: 98-108.
Reference Link - Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol 2012; 30: 488-496.
Reference Link - MacFarlane M, Harper N, Snowden RT, Dyer MJ, Barnett GA, Pringle JH et al. Mechanisms of resistance to TRAIL-induced apoptosis in primary B cell chronic lymphocytic leukaemia. Oncogene 2002; 21: 6809-6818.
Reference Link - Endo T, Nishio M, Enzler T, Cottam HB, Fukuda T, James DF et al. BAFF and APRIL support chronic lymphocytic leukemia B-cell survival through activation of the canonical NF-kB pathway. Blood 2007; 109: 703-710.
Reference Link - M. Nishikori, Classical and alternative NF-kB activation pathways and their roles in lymphoid malignancies J. Clin. Exp. Hematopathol. 45 (2005) 15-24.
Reference Link - Ihekwaba AE, Broomhead DS, Grimley RL, Benson N, Kell DB. Sensitivity analysis of parameters controlling oscillatory signalling in the NF-kappaB pathway: the roles of IKK and IkappaBalpha. Syst Biol 2001; 1(1): 93-103.
Reference Link - Nikolov S, Vera J, Rath O, Kolch W, Wolkenhauer O. Role of inhibitory proteins as modulators of oscillations in NFkB signalling. IET Syst Biol 2009; 3: 59-76.
Reference Link - Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kB dynamics reveal digital activation and analogue information processing, Nature 2010; 466: 267-271.
Reference Link - Kofler DM, Gawlik BB, Elter T, Gianella-Borradori A, Wendtner CM, Hallek M. Phase 1b trial of atacicept a recombinant protein binding BLyS and APRIL, in patients with chronic lymphocytic leukemia. Leukemia 2012; 26(4) 841-844.
Reference Link - Neychev VK, Nikolova E, Zhelev N, Mitev VI. Saponins from Tribulus terrestris L are less toxic for normal human fibroblasts than for many cancer lines: influence on apoptosis and proliferation. Exp Biol Med (Maywood) 2007; 232(1): 126-133.
- Sarek J, Klinot J, Dzubák P, Klinotová E, Nosková V, Krecek V, New lupane derived compounds with pro-apoptotic activity in cancer cells: synthesis and structure-activity relationships. J Med Chem 2003; 46(25): 5402-5415.
Reference Link