Introduction: the p53 protein
The p53 tumour suppressor protein functions principally as a tightly-regulated transcription factor that encompasses both transactivation and repression activities [
p53 also regulates genes essential for other cellular processes, often when present at basal, non-induced levels. For example, by regulating expression of leukaemia inhibitory factor (LIF), p53 is able to control implantation and hence fertility [
Structurally, p53 comprises several domains that are crucial for mediating its varied functions (Fig. 2). There are two adjoining transactivation domains, termed TAD1 and TAD2 respectively, at the N-terminus. TAD2 overlaps with a proline-rich domain and plays important roles in repression, apoptosis, and the response to gamma-irradiation [
![Figure 2. Modular structure and location of key DNA damage-induced phosphorylation sites in p53, MDM2 and MDM4. The p53, MDM2 and MDM4 proteins are shown schematically, highlighting important functional domains. In the p53 structure Boxes I-V represent the highly conserved regions of p53 first highlighted by Soussi and May [201]. Those sites of modification that are directly relevant to the DNA damage (strand break) response are shown (yellow ellipses), together with the protein kinases(s) known to phosphorylate them. The sites of acetylation (red circle) and ubiquitylation (green circle) in p53 are also indicated. Comprehensive lists of the post-translational modifications in these proteins are available elsewhere [14, 52, 62, 65, 202, 203]. Figure 2](https://biodiscovery.pensoft.net/showimg/oo_85806.jpg)
p53 is a member of a family that includes p63 and p73, both of which share a high degree of structural similarity with p53 [
The TP53 gene (encoding p53) is mutated at high frequency during the development of a wide range of cancers [
p53 and tumour suppression
It is well established that p53 provides a critical barrier to the development of many, if not all, cancer types. p53-null mice are susceptible to spontaneous tumour formation, with the elimination of wild type p53 expression in mouse models for various cancers leading to rapid tumour development and death (e.g. see [
There is also new evidence indicating that p53-dependent transcription of a large number of its responsive genes is dispensable for the suppression of some cancers and that only a small number of generally less well-characterised p53-responsive genes (but including BAX) have a crucial involvement in this process [
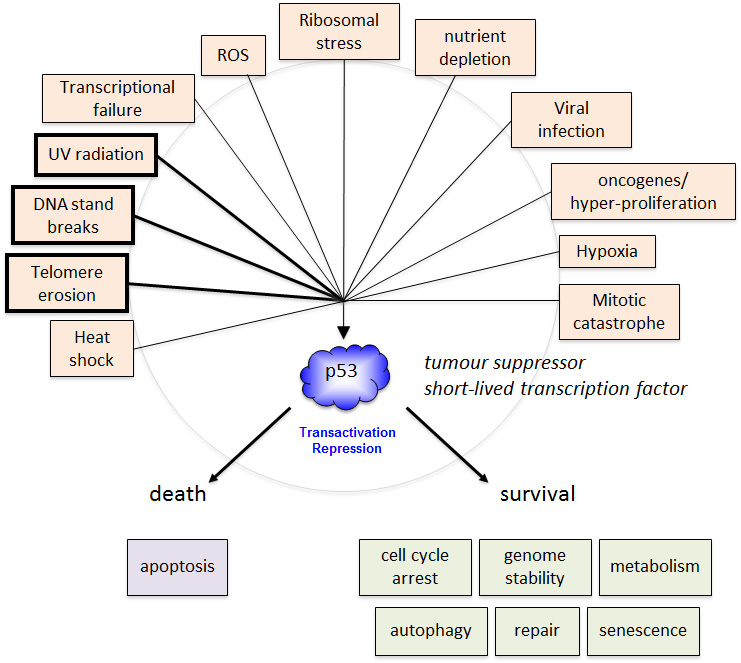
The mechanism of induction/activation of p53, and the duration of the response, may be key factors in determining whether p53 tumour suppressor function is activated. For example, developing cancer cells undergo numerous stresses including hypoxia, nutrient limitation, hyper-proliferative signalling (activated oncogenes), and persistent DNA damage. However it is still unresolved what initiating signals and/or pathways arising from such changes are primarily responsible for activating p53 tumour suppressor function. On the one hand, there is good evidence that the DNA damage pathways operate in early developing cancer cells, possibly through oncogene-driven, inappropriate activation of origins of replication, leading to replication fork collapse and strand breaks [
Regulation of p53 by MDM2
p53 is normally kept at low levels through ubiquitylation and proteasomal degradation mediated principally by the RING-finger type E3 ligase MDM2 [
The most plausible model for p53 activation is the “anti-repression” model proposed by Gu and colleagues [
MDM2 is the most extensively characterised ubiquitin ligase targeting p53 and is essentially ubiquitous in cells and tissues. It is critical for p53 regulation as exemplified by the lethality of mice lacking MDM2 expression [
Regulation of p53 by post-translational modification
In addition to ubiquitylation, the p53 protein is subject to a wide range of other post-translational modifications including multi-site phosphorylation, acetylation, methylation, and SUMOylation. Many of these have been described and discussed in detail elsewhere and will not be revisited here [
A key role for p53 acetylation
Acetylation of lysine residues is considered to be fundamentally important for activating p53 function [
In addition to the six C-terminal lysines, p53 is acetylated at three other important regulatory sites. Acetylation of K320 in the tetramerisation domain is mediated by PCAF [
In the core (site-specific DNA binding) domain, acetylation of K120 by TIP60 (KAT5)/hMOF (MYST1/KAT8) occurs rapidly after DNA damage. This modification is considered indispensable for the activation of p53 target genes encoding apoptosis-associated proteins, but thought to have little influence on the expression of genes encoding proteins required for cell cycle arrest [
The role of these lysines in vivo has been investigated using knock-in mouse models in which either six C-terminal lysines (K367, K369, K370, K378, K379, and K383, equivalent to human K370, K372, K373, K381, K382 and K386 respectively), or these six C-terminal lysines plus K384 (murine specific), are substituted by arginine (p536KR and p537KR respectively). In both cases, however, the mutant mice showed only mild phenotypes and generally showed no major differences in growth arrest, apoptosis or tumour suppression [
p53 induction and activation
The main event in the induction of p53 is the uncoupling of p53 from degradation, mediated by MDM2 [
Induction of p53 in response to activated oncogenes
Activated oncogenes (such as Ras, Myc, E2F-1, beta-catenin) use various mechanisms to drive up the levels of ARF, an inhibitor of MDM2 which is encoded by the gene CDKN2A and overlaps with the INK4A locus. These mechanisms include stimulation of ARF transcription, inhibition of ARF degradation and segregation of ARF from its targeting ubiquitin ligase, ULF (reviewed in [
Induction of p53 in response to ribosomal stress
A different mechanism of p53 induction is employed in response to ribosomal stress (also known as nucleolar stress), which arises when the highly coordinated process of ribosome synthesis and assembly is disrupted [
Induction of p53 by MDM2-targeted drugs
The development of drugs which induce p53 offer significant insight into the mechanisms of activation and their downstream effects. Perhaps the best characterised of these drugs is Nutlin-3a (hereafter Nutlin) which competes with the binding of p53 to the N-terminus of MDM2 [
Other mechanisms can influence the potency of these pathways. For example, survival signalling and/or oncogenic signalling via activated AKT can stimulate MDM2 activity and increase the threshold needed to induce a p53 response [
The DNA damage pathways: induction, activation and the importance of phosphorylation
The induction and activation of p53 in response to DNA damage is orchestrated by the ATM and ATR protein kinases which are activated by double- and single-strand breaks respectively. A major part of this response involves the coordination and integration of a number of signalling pathways leading to changes in the post-translational status of p53 itself and several of its direct or indirect regulators [
Phosphorylation of the N-terminus of p53 inhibits MDM2 association and stimulates interaction with transcription factors
In response to double strand breaks, the activation and phosphorylation of p53 are rapid events that occur within the first 30 minutes following the stimulus. ATM activation can be transient (lasting only a few hours) and is succeeded, in an overlapping manner, by the activation of ATR, possibly through the generation of single stranded stretches of DNA that are generated by the repair responses. ATM and ATR both phosphorylate Serine 15 in p53 (Fig. 2): thus, the consecutive activation of these two protein kinases provides a continuity of p53 phosphorylation that endures for several hours after the initial stimulus.
Phosphorylation of Ser15 is considered to be an initiating and nucleating event in p53 activation [
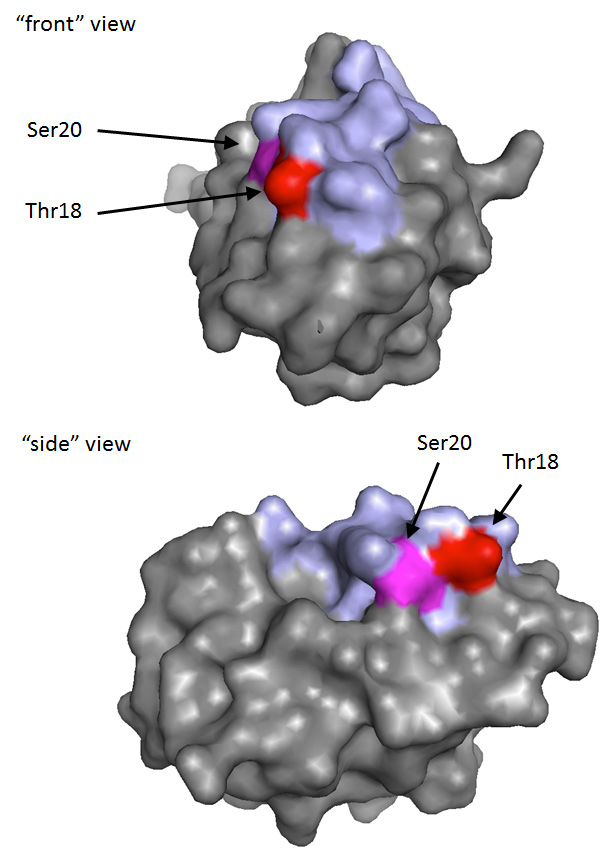
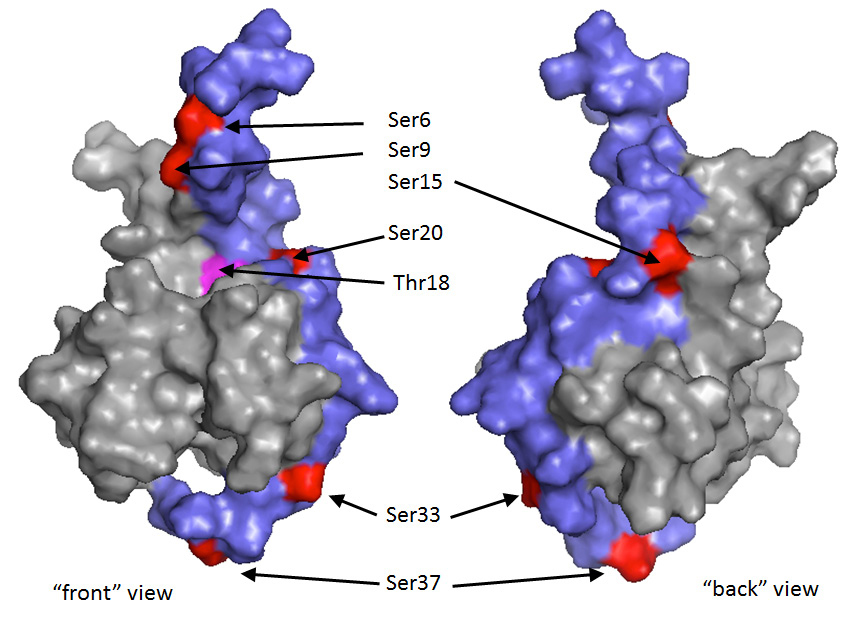
Other residues in the TAD1 region of p53 are also targeted for DNA damage-induced phosphorylation, including serines 6, 9, 33 and 37 [
Multiple phosphorylation events regulate transcription factor/co-activator recruitment and may act cooperatively as a “rheostat” for stimulating p53 activity
In addition to blocking the interaction with MDM2, phosphorylation of individual residues in the N-terminus (TAD1) of p53 stimulates, to various degrees, interaction with p53 binding sites in the p300 and CBP transcriptional co-activator proteins (reviewed in detail previously in [
Additional phosphorylation events in the TAD1 region of p53 (serines 6, 9, 33 and 37: mentioned above) may have an important role. Combinations of the various TAD1 phosphorylation events have been demonstrated to act cooperatively in stimulating p300/CBP binding in a graded or incremental manner [
Taken together, the above studies establish two important principles in regulating p53 induction and activation: firstly, that phosphorylation of these key N-terminal sites acts as a switch in which rapid uncoupling of MDM2 and recruitment of key transcription factors can occur; secondly that cooperation between the different phosphorylation sites may act as a rheostat to permit fine-tuning of the association between p53 and p300/CBP.
Phosphorylations of other key sites in p53 have a major impact on the response to DNA damage and mediate specific interactions with different transcriptional proteins
Other sites in p53 are modified in response to DNA damage, in some cases sequentially depending on events at the N-terminus. Of particular interest, Ser46 in the TAD2 domain is phosphorylated in a manner that may be dependent, directly or indirectly, upon activation of the ATM pathway [
Several other well-characterised phosphorylation sites in p53 can contribute to the DNA damage response [
Subtle or even extensive variations in the extent of phosphorylation and other forms of modification of p53 residues occur depending on the inducing stimulus, which governs the type of DNA damage acquired, and the intensity and duration of the stimulus (e.g. see [
The role of phosphorylation of p53 regulators in p53 induction by DNA damage
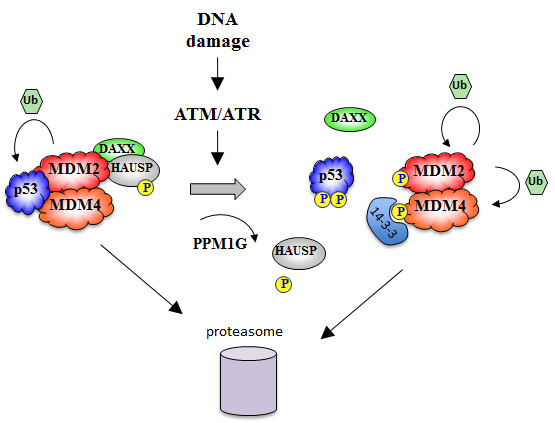
Post-translational events in p53 are part of a broader mechanism of induction and activation that involves simultaneous modification of other p53 regulators including MDM2 and MDM4 (Fig. 2). MDM2 and p53 interact tightly through several points of contact that form the targets of inducing signals. A hydrophobic cleft in the N-terminus of MDM2 serves as a docking site for three key hydrophobic residues in the N-terminus of p53: F19, W23 and L26. Association of p53 and MDM2 through this high affinity interaction is thought to lead to a conformational shift that stimulates an essential low affinity contact between the central acidic domain of MDM2 and the BoxIV/V region of p53 [
In response to strand breaks, ATM phosphorylates MDM2 at various C-terminal sites. Initially this response was thought to be focused on Ser395 [
Confirmation that ATM-mediated phosphorylation of MDM2 plays a critical role in the induction of p53 in vivo was provided recently following the generation of Mdm2S394A/S394A and Mdm2S394D/S394D mice [
The acidic domain of MDM2 contains a cluster of residues that are phosphorylated under normal unstressed conditions and, based on mutational analyses, contribute towards the normal turnover of p53 [
MDM4 is a defective E3 ligase that is structurally very similar to MDM2 (Fig. 2). Like, MDM2, MDM4 is also targeted by the DNA damage pathways, leading to changes in its phosphorylation status. In response to DNA damage, Ser342 and Ser367 in MDM4 are phosphorylated by CHK2 while Ser403 is directly phosphorylated by ATM [
Similar to MDM2, MDM4 is also phosphorylated by c-ABL in response to DNA damage. The modified residues are Tyr55 and Tyr99, both of which are located in the p53 binding domain. Phosphorylation of Tyr99 impairs p53 binding, thereby facilitating p53 activation [
The phosphorylation of an isoform of HAUSP (USP7S) also contributes to the DNA damage-mediated induction of p53 [
Taken together, these various studies indicate that DNA damage gives rise to a highly coordinated and integrated process part of which involves the targeting of several interacting proteins involved in promoting p53 turnover (together with many targets in the DNA repair machinery) through protein phosphorylation mechanisms. These events constitute an integrated response in which p53 induction and activation are tailored in accordance with the type, intensity and duration of the initiating stimulus.
Attenuation of p53 induction by DNA damage
Once induced, activated p53 mediates the expression of key negative regulators, mainly MDM2 and the WIP1 phosphatase [
The WIP1 (PPM1D) phosphatase is also induced by p53 and underpins this response [
Mouse models expressing phosphorylation site mutants of p53 and its regulatory partners: what they tell us
A number of studies have carefully considered the biological relevance of DNA damage-induced phosphorylation events in p53 and its principal regulators, MDM2 and MDM4, in vivo, by generating knock-in mice bearing amino acid substitutions at one or more of these key positions (
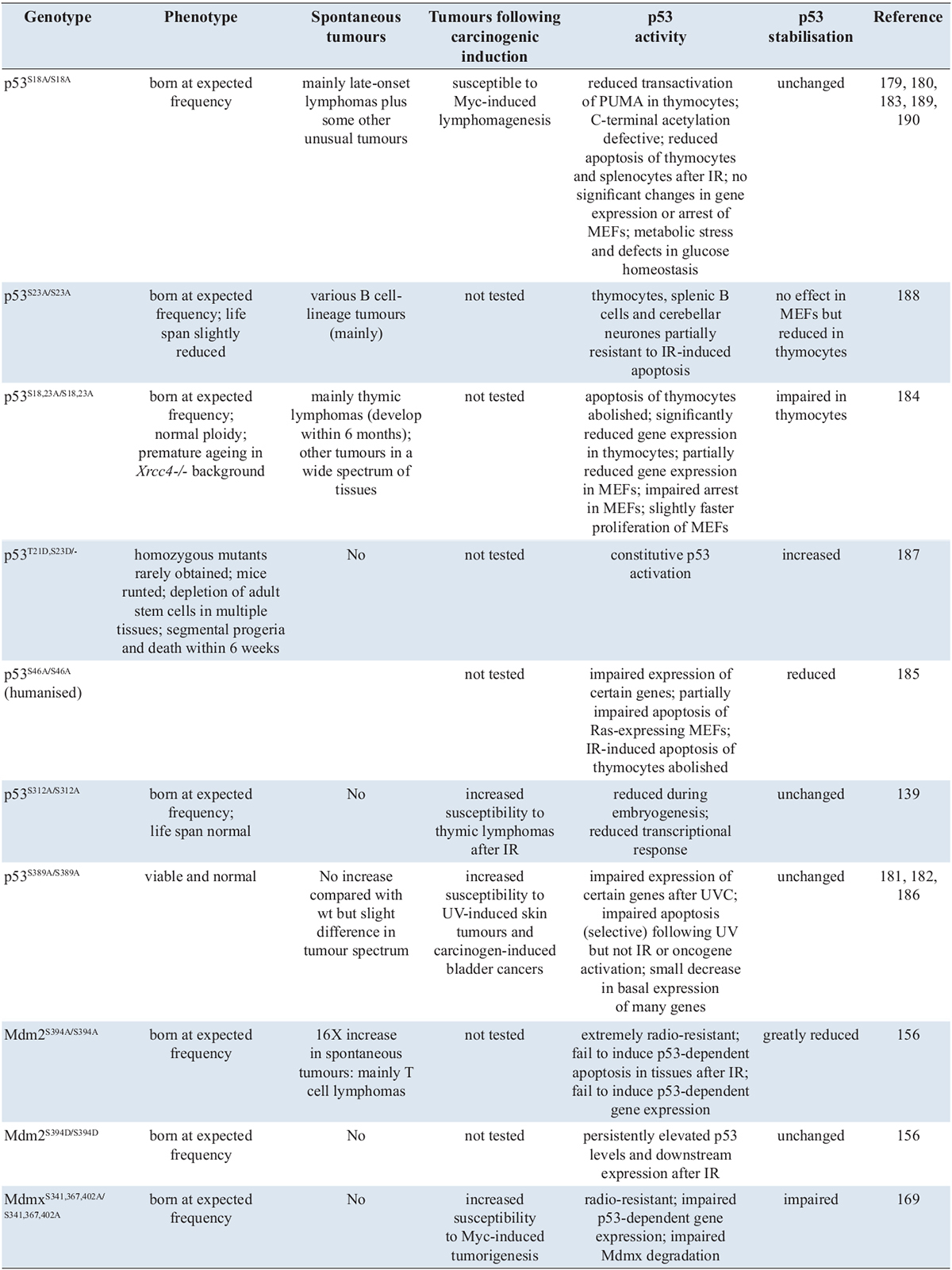
(1) The p53S18A/S18A mouse (murine Ser18 is equivalent to human Ser15) was the first to be generated and is the most consistently and intensively studied [
(2) There is cooperation between different modification sites in vivo, reflecting the model described above based on biochemical and cultured cell analyses. This is perhaps best exemplified by the p53S18,23A/S18/23A mouse [
(3) While the mice collectively demonstrate that phosphorylation is not essential for tumour suppression, they clearly show that phosphorylation contributes, probably in a cell- or tissue-dependent manner, to inhibiting cancer development.
(4) The effects of substituting phosphorylation sites on p53 function in cells of lymphoid lineage are more acute as compared with those in fibroblasts. These differences highlight the possibility that phosphorylation of p53 may play fundamental roles in certain cells types but may be redundant or even irrelevant in others. It could be argued that such differences in cell behaviour could reflect artifactual changes acquired upon isolation of the cells and growth in culture. However, the increased sensitivity to lymphoma development in several of the mice tends to argue against this and may therefore support lineage-dependent behaviour.
(5) Individual phosphorylation sites in p53 may have highly specific and/or cell type-dependent roles in vivo. For example, the S389A mice show a striking selectivity in gene expression and in contributing to tumour inhibition [
(6) While individual, or combinations of, phosphorylation sites in p53 have subtle and cell specific roles, the targeting of MDM2 via the DNA damage pathways (via Ser394 phosphorylation) has a major effect on the ability to induce and activate p53. This dominance of MDM2 fits well with the growing understanding that uncoupling the p53/MDM2 interaction is the key principle underlying p53 activation [
(7) In the growing list of biological functions of p53, its ability to coordinately regulate the levels of key metabolic enzymes has acquired enormous significance, both in a physiological context and in the context of cancer prevention by opposing the Warburg effect (aerobic glycolysis in cancer cells) [
Perspectives
The wealth of study on the role of p53 phosphorylation discussed above raises a number of interesting issues for further consideration and exploration.
What is the role of phosphorylation?
Recent evidence has underpinned the idea that, while DNA damage signals impinging on p53 lead to multiple phosphorylation events on key players including MDM2, MDM4 and p53 itself, the targeting of MDM4 [
An alternative possibility is that the efficiency of this induction process is improved by the sort of coordinate control that multi-site phosphorylation applies to MDM2, p53 and other proteins involved in regulating p53 levels: e.g. inactivating MDM2 and MDM4 while simultaneously blocking their interaction sites on p53 could ensure the outcome and increase the strength of the induction. Moreover, if induction occurs via the “anti-repression” model [
What does phosphorylation do on chromatin?
Related to points raised in the previous paragraph is the issue of the role played by multi-site phosphorylation in the context of chromatin where p53 conducts most of its business. It is clear that phosphorylation can block MDM2 interaction and promote recruitment of key transcription factors. However, other (DNA damage-independent) stresses are also able to recruit transcription factors so is there a fundamental requirement for phosphorylation in this process? This seems unlikely given that other stress stimuli do not induce these modifications on p53. However, it may be that DNA damage is a special case: thus, for example, if DNA damage-mediated modification of other transcriptional components occurs, (or, indeed recruitment of different transcription factors), p53 phosphorylation may be required to be compatible with these. Or perhaps selectivity in the strength of inducing certain genes is required (i.e. turning them up or down as compared with other stresses as opposed to on or off). Given that small changes in p53 activity can have major impacts on cell fate, the value of subtle levels of regulation cannot be over-emphasised.
What p53-dependent biological outcomes are actually regulated?
The evidence discussed above, particularly that from the animal models, indicates that phosphorylation of p53 itself is likely to have a bearing on the ability of p53 to mediate tumour suppression (Table 1). However, these data collectively indicate that such a role is contributory as opposed to fundamental and may actually have cell- or tissue-type relevance. If phosphorylation of p53 is not essential for tumour suppression, then what function(s) has it evolved to regulate? One possibility is stem cell status where, for example, Ser315 phosphorylation appears to play and important role in controlling nanog expression [
Is there a role for phosphorylation of mutant p53?
Mutant p53 proteins are now known to help drive tumour progression and metastasis. Additionally, it is becoming clear that many of the drugs used in the clinic can activate the cancer-promoting functions of mutant p53 [
References
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harbor Perspect Biol 2010; 2: a000935.
Reference Link - Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harbor Perspect Biol 2010; 2: a000893.
Reference Link - Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402-412.
Reference Link - Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009; 137: 413-431.
Reference Link - Hu W, Feng Z, Levine AJ. The Regulation of Multiple p53 Stress Responses is Mediated through MDM2. Genes Cancer 2012; 3: 199-208.
Reference Link - Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol 2007; 8: 275-283.
Reference Link - Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol 2011; 12: 259-265.
Reference Link - Solozobova V, Blattner C. p53 in stem cells. World J Biol Chem 2011; 2: 202-214.
Reference Link - Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev 2004; 18: 306-319.
Reference Link - Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002; 415: 45-53.
Reference Link - Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol 2010; 20: 427-434.
Reference Link - Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer 2009; 9: 691-700.
Reference Link - Campbell HG, Mehta R, Neumann AA, Rubio C, Baird M, Slatter TL, et al. Activation of p53 following ionizing radiation, but not other stressors, is dependent on the proline-rich domain (PRD). Oncogene 2013; 32: 827-836.
Reference Link - Anderson CW, Appella E. Signaling to the p53 tumor suppressor through pathways activated by genotoxic and non-genotoxic stresses. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling: Elsevier; 2009. p. 237-247.
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 2001; 268: 2764-2772.
Reference Link - Carr SM, Munro S, La Thangue NB. Lysine methylation and the regulation of p53. Essays Biochem 2012; 52: 79-92.
- Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med 2010; 16: 528-536.
Reference Link - Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harbor Perspect Biol 2010; 2: a004887.
Reference Link - Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 2005; 7: 363-373.
Reference Link - Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ 2011; 18: 1815-1824.
Reference Link - Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev 2012; 26: 1268-1286.
Reference Link - Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol 2011; 223: 116-126.
Reference Link - Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005; 436: 725-730.
Reference Link - Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 2007; 21: 379-384.
Reference Link - Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 1999; 13: 2658-2669.
Reference Link - Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 2006; 127: 1323-1334.
Reference Link - Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007; 445: 661-665.
Reference Link - Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007; 445: 656-660.
Reference Link - Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012; 149: 1269-1283.
Reference Link - Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 2011; 145: 571-583.
Reference Link - Jiang D, Brady CA, Johnson TM, Lee EY, Park EJ, Scott MP, et al. Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proc Natl Acad Sci U S A 2011; 108: 17123-17128.
Reference Link - Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006; 444: 633-637.
Reference Link - Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006; 444: 638-642.
Reference Link - Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 1999; 13: 2670-2677.
Reference Link - Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 2006; 443: 214-217.
Reference Link - Hinkal G, Parikh N, Donehower LA. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS One 2009; 4: e6654.
Reference Link - Efeyan A, Garcia-Cao I, Herranz D, Velasco-Miguel S, Serrano M. Tumour biology: Policing of oncogene activity by p53. Nature 2006; 443: 159.
Reference Link - Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, et al. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One 2009; 4: e5475.
Reference Link - Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell 2006; 21: 307-315.
Reference Link - Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995; 378: 206-208.
Reference Link - Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378: 203-206.
Reference Link - Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 2006; 10: 501-514.
Reference Link - Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome- mediated degradation. Mol Cell Biol 2000; 20: 8458-8467.
Reference Link - Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at p53 C terminus in regulating p53 stability and activity. Mol Cell Biol 2005; 25: 5389-5395.
Reference Link - Krummel KA, Lee CJ, Toledo F, Wahl G. The C-terminal lysines fine-tune p53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A 2005; 102: 10188-10193.
Reference Link - Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003; 302: 1972-1975.
Reference Link - Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993; 7: 1126-1132.
Reference Link - Kruse JP, Gu W. Modes of p53 regulation. Cell 2009; 137: 609-622.
Reference Link - Biderman L, Manley JL, Prives C. Mdm2 and MdmX as Regulators of Gene Expression. Genes Cancer 2012; 3: 264-273.
Reference Link - Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci 2007; 120: 371-378.
Reference Link - Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol 2007; 39: 1476-1482.
Reference Link - Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83-96.
Reference Link - Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res 2007; 67: 6026-6030.
Reference Link - Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem 2011; 286: 23725-23734.
Reference Link - Cross B, Chen L, Cheng Q, Li B, Yuan ZM, Chen J. Inhibition of p53 DNA binding function by the MDM2 protein acidic domain. J Biol Chem 2011; 286: 16018-16029.
Reference Link - Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol 2007; 9: 428-435.
Reference Link - Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH. C-terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol 2001; 21: 8521-8532.
Reference Link - Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 2008; 32: 180-189.
Reference Link - Love IM, Grossman SR. It Takes 15 to Tango: Making Sense of the Many Ubiquitin Ligases of p53. Genes Cancer 2012; 3: 249-263.
Reference Link - Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene 2005; 24: 2899-2908.
Reference Link - Bouska A, Eischen CM. Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem Sci 2009; 34: 279-286.
Reference Link - Jenkins LM, Durell SR, Mazur SJ, Appella E. p53 N-terminal phosphorylation: a defining layer of complex regulation. Carcinogenesis 2012; 33: 1441-1449.
Reference Link - MacLaine NJ, Hupp TR. How phosphorylation controls p53. Cell Cycle 2011; 10: 916-921.
Reference Link - Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 2009; 9: 714-723.
- Meek DW, Anderson CW. Posttranslational Modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol 2009; 1: a000950.
Reference Link - Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell 2008; 133: 612-626.
Reference Link - Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 2002; 277: 50607-50611.
Reference Link - Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol 1999; 19: 1202-1209.
- Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 2006; 173: 533-544.
Reference Link - Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, et al. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol Cell Biol 2006; 26: 6859-6869.
Reference Link - Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 2006; 24: 841-851.
Reference Link - Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 2006; 24: 827-839.
Reference Link - Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 2005; 24: 3411-3422.
Reference Link - Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 2000; 275: 8945-8951.
Reference Link - Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 2000; 19: 1473-1476.
Reference Link - Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A 2005; 102: 5056-5061.
Reference Link - Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, et al. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol 2007; 9: 331-338.
Reference Link - Feng J, Tamaskovic R, Yang Z, Brazil DP, Merlo A, Hess D, et al. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem 2004; 279: 35510-35517.
Reference Link - Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A 2006; 103: 3232-3237.
Reference Link - Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 2004; 13: 879-886.
Reference Link - Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 2005; 18: 565-576.
Reference Link - Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol 2006; 8: 855-862.
Reference Link - Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell 2006; 9: 273-285.
Reference Link - Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006; 6: 909-923.
Reference Link - Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol 2006; 13: 285-291.
Reference Link - Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget 2011; 2: 234-238.
- Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene 2004; 23: 3217-3221.
Reference Link - Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell 2010; 18: 231-243.
Reference Link - Pan W, Issaq S, Zhang Y. The in vivo role of the RP-Mdm2-p53 pathway in signaling oncogenic stress induced by pRb inactivation and Ras overexpression. PLoS One 2011; 6: e21625.
Reference Link - Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol 2009; 49: 223-241.
Reference Link - Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844-848.
Reference Link - Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A 2001; 98: 11598-11603.
Reference Link - Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 2001; 3: 973-982.
Reference Link - Feng Z, Liu L, Zhang C, Zheng T, Wang J, Lin M, et al. Chronic restraint stress attenuates p53 function and promotes tumorigenesis. Proc Natl Acad Sci U S A 2012; 109: 7013-7018.
Reference Link - Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 2004; 119: 591-602.
Reference Link - Saito S, Goodarzi AA, Higashimoto Y, Noda Y, Lees-Miller SP, Appella E, et al. ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J Biol Chem 2002; 277: 12491-12494.
Reference Link - Craig AL, Burch L, Vojtesek B, Mitukovska J, Thompson A, R. HT. Novel phosphorylation sites of human tumour suppressor protein p53 at ser20 and thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem J 1999; 342: 133-141.
Reference Link - Dumaz N, Milne DM, Meek DW. Protein kinase CK1 is a p53 threonine-18 protein kinase which requires prior phosphorylation of serine 15. FEBS Lett 1999; 463: 312-316.
Reference Link - Feng H, Jenkins LM, Durell SR, Hayashi R, Mazur SJ, Cherry S, et al. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure 2009; 17: 202-210.
Reference Link - Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci U S A 2009; 106: 6591-6596.
Reference Link - Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci U S A 2010; 107: 19290-19295.
Reference Link - Sakaguchi K, Saito S, Higashimoto Y, Roy S, Anderson CW, Appella E. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem 2000; 275: 9278-9283.
Reference Link - Teufel DP, Bycroft M, Fersht AR. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene 2009; 28: 2112-2118.
Reference Link - Brown CJ, Srinivasan D, Jun LH, Coomber D, Verma CS, Lane DP. The electrostatic surface of MDM2 modulates the specificity of its interaction with phosphorylated and unphosphorylated p53 peptides. Cell Cycle 2008; 7: 608-610.
Reference Link - Lee HS, Hauber SO, Vindus D, Niemeyer M. Isostructural potassium and thallium salts of sterically crowded triazenes: a structural and computational study. Inorg Chem 2008; 47: 4401-4412.
Reference Link - Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 2001; 292: 1910-1915.
Reference Link - Craig AL, Chrystal JA, Fraser JA, Sphyris N, Lin Y, Harrison BJ, et al. The MDM2 ubiquitination signal in the DNA-binding domain of p53 forms a docking site for calcium calmodulin kinase superfamily members. Mol Cell Biol 2007; 27: 3542-3555.
Reference Link - Dumaz N, Milne DM, Jardine LJ, Meek DW. Critical roles for the serine 20, but not the serine 15, phosphorylation site and for the polyproline domain in regulating p53 turnover. Biochem J 2001; 359: 459-464.
Reference Link - Jabbur JR, Zhang W. p53 Antiproliferative function is enhanced by aspartate substitution at threonine 18 and serine 20. Cancer Biol Ther 2002; 1: 277-283.
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 1998; 12: 2831-2841.
Reference Link - Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation domains of p53. Proc Natl Acad Sci U S A 2007; 104: 7009-7014.
Reference Link - Jenkins LM, Yamaguchi H, Hayashi R, Cherry S, Tropea JE, Miller M, et al. Two Distinct Motifs within the p53 Transactivation Domain Bind to the Taz2 Domain of p300 and Are Differentially Affected by Phosphorylation. Biochemistry 2009; 48: 1244-1255.
Reference Link - Lee CW, Martinez-Yamout MA, Dyson HJ, Wright PE. Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry 2010; 49: 9964-9971.
Reference Link - D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002; 4: 11-19.
Reference Link - Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 2002; 4: 1-10.
Reference Link - Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 2007; 25: 725-738.
Reference Link - Yoshida K, Liu H, Miki Y. Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J Biol Chem 2006; 281: 5734-5740.
Reference Link - Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S, et al. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem 2008; 283: 3979-3987.
Reference Link - Kodama M, Otsubo C, Hirota T, Yokota J, Enari M, Taya Y. Requirement of ATM for rapid p53 phosphorylation at Ser46 without Ser/Thr-Gln sequences. Mol Cell Biol 2010; 30: 1620-1633.
Reference Link - Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102: 849-862.
Reference Link - Mayo LD, Seo YR, Jackson MW, Smith ML, Rivera Guzman J, Korgaonkar CK, et al. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J Biol Chem 2005; 280: 25953-25959.
Reference Link - Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, et al. Regulation of PTEN transcription by p53. Mol Cell 2001; 8: 317-325.
Reference Link - Smeenk L, van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG, et al. Role of p53 serine 46 in p53 target gene regulation. PLoS One 2011;6: e17574.
Reference Link - Cai X, Liu X. Inhibition of Thr-55 phosphorylation restores p53 nuclear localization and sensitizes cancer cells to DNA damage. Proc Natl Acad Sci U S A 2008; 105: 16958-16963.
Reference Link - Li HH, Li AG, Sheppard HM, Liu X. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: a role for TAF1 in cell G1 progression. Mol Cell 2004; 13: 867-878.
Reference Link - Shouse GP, Cai X, Liu X. Serine 15 phosphorylation of p53 directs its interaction with B56gamma and the tumor suppressor activity of B56gamma-specific protein phosphatase 2A. Mol Cell Biol 2008; 28: 448-456.
Reference Link - Di Lello P, Jenkins LMM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, et al. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell 2006; 22: 731-740.
Reference Link - Di Lello P, Miller Jenkins LM, Mas C, Langlois C, Malitskaya E, Fradet-Turcotte A, et al. p53 and TFIIEalpha share a common binding site on the Tfb1/p62 subunit of TFIIH. Proc Natl Acad Sci U S A 2008; 105: 106-111.
Reference Link - Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, et al. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science 2007; 315: 840-843.
Reference Link - Knippschild U, Milne DM, Campbell LE, DeMaggio AJ, Christenson E, Hoekstra MF, et al. p53 is phosphorylated in vitro and in vivo by the delta and epsilon isoforms of casein kinase 1 and enhances the level of casein kinase 1 delta in response to topoisomerase-directed drugs. Oncogene 1997; 15: 1727-1736.
Reference Link - Saito S, Yamaguchi H, Higashimoto Y, Chao C, Xu Y, Fornace Jr AJ, et al. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J Biol Chem 2003; 278: 37536-37544.
Reference Link - Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, et al. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J 1999; 18: 6845-6854.
Reference Link - Cox ML, Meek DW. Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell Signal 2010; 22: 564-571.
Reference Link - Serrano MA, Li Z, Dangeti M, Musich PR, Patrick S, Roginskaya M, et al. DNA-PK, ATM and ATR collaboratively regulate p53-RPA interaction to facilitate homologous recombination DNA repair. Oncogene 2012; doi:10.1038/onc.2012.257
Reference Link - Berger M, Stahl N, Del Sal G, Haupt Y. Mutations in proline 82 of p53 impair its activation by Pin1 and Chk2 in response to DNA damage. Mol Cell Biol 2005; 25: 5380-5388.
Reference Link - Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J 2001; 20: 1630-1639.
Reference Link - Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, et al. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem 2004; 279: 52175-52182.
Reference Link - Blaydes JP, Luciani MG, Pospisilova S, Ball HM-L, Vojtesek B, Hupp TR. Stoichiometric phosphorylation of human p53 at Ser315 stimulates p53-dependent transcription. J Biol Chem 2001; 14: 4699-4708.
Reference Link - Slee EA, Benassi B, Goldin R, Zhong S, Ratnayaka I, Blandino G, et al. Phosphorylation of Ser312 contributes to tumor suppression by p53 in vivo. Proc Natl Acad Sci U S A 2010; 107: 19479-19484.
Reference Link - Fraser JA, Madhumalar A, Blackburn E, Bramham J, Walkinshaw MD, Verma C, et al. A novel p53 phosphorylation site within the MDM2 ubiquitination signal: II. a model in which phosphorylation at SER269 induces a mutant conformation to p53. J Biol Chem 2010; 285: 37773-37786.
Reference Link - Fraser JA, Vojtesek B, Hupp TR. A novel p53 phosphorylation site within the MDM2 ubiquitination signal: I. phosphorylation at SER269 in vivo is linked to inactivation of p53 function. J Biol Chem 2010; 285: 37762-37772.
Reference Link - Waterman MJF, Stavridi ES, F. WJL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet 1998; 19: 175-178.
Reference Link - Latonen L, Taya Y, Laiho M. UV-radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene 2001; 20: 6784-6793.
Reference Link - Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene 2008; 27: 4013-4023.
Reference Link - Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol 2008; 9: 702-712.
Reference Link - Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem 2006; 281: 28575-28583.
Reference Link - Shimizu H, Burch LR, Smith AJ, Dornan D, Wallace M, Ball KL, et al. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J Biol Chem 2002; 277: 28446-28458.
Reference Link - Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell 2006; 23: 251-263.
Reference Link - Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci U S A 2006; 103: 1227-1232.
Reference Link - Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001; 15: 1067-1077.
Reference Link - Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J 2009; 28: 3857-3867.
Reference Link - Shinozaki T, Nota A, Taya Y, Okamoto K. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene 2003; 22: 8870-8880.
Reference Link - Cheng Q, Cross B, Li B, Chen L, Li Z, Chen J. Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol Cell Biol 2011; 31: 4951-4963.
Reference Link - Goldberg Z, Sionov RV, Berger M, Zwang Y, Perets R, Van Etten RA, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J 2002; 21: 3715-3727.
Reference Link - Li YC, Wahl GM. What a difference a phosphate makes: life or death decided by a single amino acid in MDM2. Cancer Cell 2012; 21: 595-596.
Reference Link - Gannon HS, Woda BA, Jones SN. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell 2012; 21: 668-679.
Reference Link - Blattner C, Hay TJ, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol 2002; 22: 6170-6182.
Reference Link - Hay TJ, Meek DW. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett 2000; 478: 183-186.
Reference Link - Hjerrild M, Milne D, Dumaz N, Hay T, Issinger OG, Meek D. Phosphorylation of murine double minute clone 2 (MDM2) protein at serine-267 by protein kinase CK2 in vitro and in cultured cells. Biochem J 2001; 355: 347-356.
Reference Link - Boehme KA, Kulikov R, Blattner C. p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc Natl Acad Sci U S A 2008; 105: 7785-7790.
Reference Link - Kulikov R, Boehme KA, Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol Cell Biol 2005; 25: 7170-7180.
Reference Link - Winter M, Milne D, Dias S, Kulikov R, Knippschild U, Blattner C, et al. Protein kinase CK1-delta phosphorylates key sites in the acidic domain of Mdm2 that regulate p53 turnover. Biochemistry 2004; 43: 16356-16364.
Reference Link - Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell 2008; 30: 203-213.
Reference Link - Surucu B, Bozulic L, Hynx D, Parcellier A, Hemmings BA. In vivo analysis of PKB/Akt regulation in DNA-PKcs-null mice reveals a role for PKB/Akt in DNA damage response and tumorigenesis. J Biol Chem 2008; 283: 30025-30033.
Reference Link - Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, et al. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J 2006; 25: 1207-1218.
Reference Link - Pereg Y, Lam S, Teunisse AF, Biton S, Meulmeester E, Mittelman L, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol 2006; 26: 6819-6831.
Reference Link - LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J 2006; 25: 1196-1206.
Reference Link - Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 2005; 25: 9608-9620.
Reference Link - Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell 2009; 16: 33-43.
Reference Link - Zuckerman V, Lenos K, Popowicz GM, Silberman I, Grossman T, Marine JC, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem 2009; 284: 4031-4039.
Reference Link - Khoronenkova SV, Dianova, II, Ternette N, Kessler BM, Parsons JL, Dianov GL. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell 2012; 45: 801-813.
Reference Link - Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A 1997; 94: 6048-6053.
Reference Link - Rossi M, Demidov ON, Anderson CW, Appella E, Mazur SJ. Induction of PPM1D following DNA-damaging treatments through a conserved p53 response element coincides with a shift in the use of transcription initiation sites. Nucleic Acids Res 2008; 36: 7168-7180.
Reference Link - Guo L, Liew HP, Camus S, Goh AM, Chee LL, Lunny DP, et al. Ionizing radiation induces a dramatic persistence of p53 protein accumulation and DNA damage signaling in mutant p53 zebrafish. Oncogene 2012; doi: 10.1038/onc.2012.409
Reference Link - Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 2008; 22: 1337-1344.
Reference Link - Le Guezennec X, Bulavin DV. WIP1 phosphatase at the crossroads of cancer and aging. Trends Biochem Sci 2010; 35: 109-114.
Reference Link - Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev 2005; 19: 1162-1174.
Reference Link - Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 2007; 12: 342-354.
Reference Link - Armata HL, Garlick DS, Sluss HK. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Res 2007; 67: 11696-11703.
Reference Link - Armata HL, Golebiowski D, Jung DY, Ko HJ, Kim JK, Sluss HK. Requirement of the ATM/p53 tumor suppressor pathway for glucose homeostasis. Mol Cell Biol 2010; 30: 5787-5794.
Reference Link - Bruins W, Bruning O, Jonker MJ, Zwart E, van der Hoeven TV, Pennings JL, et al. The absence of Ser389 phosphorylation in p53 affects the basal gene expression level of many p53-dependent genes and alters the biphasic response to UV exposure in mouse embryonic fibroblasts. Mol Cell Biol 2008; 28: 1974-1987.
Reference Link - Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB, et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol 2004; 24: 8884-8894.
Reference Link - Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem 2003; 278: 41028-41033.
Reference Link - Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J 2006; 25: 2615-2622.
- Feng L, Hollstein M, Xu Y. Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle 2006; 5: 2812-2819.
Reference Link - Hoogervorst EM, Bruins W, Zwart E, van Oostrom CT, van den Aardweg GJ, Beems RB, et al. Lack of p53 Ser389 phosphorylation predisposes mice to develop 2-acetylaminofluorene-induced bladder tumors but not ionizing radiation-induced lymphomas. Cancer Res 2005; 65: 3610-3616.
Reference Link - Liu D, Ou L, Clemenson GD, Jr., Chao C, Lutske ME, Zambetti GP, et al. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol 2010; 12: 993-998.
Reference Link - MacPherson D, Kim J, Kim T, Rhee BK, Van Oostrom CT, DiTullio RA, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J 2004; 23: 3689-3699.
Reference Link - Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol 2004; 24: 976-984.
Reference Link - Sluss HK, Gannon H, Coles AH, Shen Q, Eischen CM, Jones SN. Phosphorylation of p53 serine 18 upregulates apoptosis to suppress Myc-induced tumorigenesis. Mol Cancer Res 2010; 8: 216-222.
Reference Link - Armata HL, Shroff P, Garlick DE, Penta K, Tapper AR, Sluss HK. Loss of p53 Ser18 and Atm results in embryonic lethality without cooperation in tumorigenesis. PLoS One 2011; 6: e24813.
Reference Link - Maddocks OD, Vousden KH. Metabolic regulation by p53. J Mol Med 2011; 89: 237-245.
Reference Link - d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003; 426: 194-198.
Reference Link - Khalil HS, Tummala H, Zhelev N. ATM in focus: a damage sensor and cancer target. Biodiscovery 2012; 5: 1.
- Webley K, Bond JA, Jones CJ, Blaydes JP, Craig A, Hupp T, et al. Posttranslational modifications of p53 in replicative senescence overlapping but distinct from those induced by DNA damage. Mol Cell Biol 2000; 20: 2803-2808.
Reference Link - Yang C, Tang X, Guo X, Niikura Y, Kitagawa K, Cui K, et al. Aurora-B mediated ATM serine 1403 phosphorylation is required for mitotic ATM activation and the spindle checkpoint. Mol Cell 2011;44: 597-608.
Reference Link - Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 2011; 43: 117-120.
Reference Link - Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7: 165-171.
Reference Link - Meek DW. The role of p53 in the response to mitotic spindle damage. Pathol Biol (Paris) 2000; 48: 246-254.
- Suh YA, Post SM, Elizondo-Fraire AC, Maccio DR, Jackson JG, El-Naggar AK, et al. Multiple stress signals activate mutant p53 in vivo. Cancer Res 2011; 71: 7168-7175.
Reference Link - Soussi T, Caron de Fromentel C, May P. Structural aspects of the p53 protein in relation to gene evolution. Oncogene 1990; 5: 945-952.
- Chen J. The Roles of MDM2 and MDMX Phosphorylation in Stress Signaling to p53. Genes Cancer 2012; 3: 274-282.
Reference Link - Meek DW, Hupp TR. The regulation of MDM2 by multisite phosphorylation-Opportunities for molecular-based intervention to target tumours? Semin Cancer Biol 2010; 20: 19-28.
Reference Link