Background
During interphase, chromosomes occupy non-random positions within the nuclear space. Interphase chromosome arrangement is considered to play significant role in development and disease mediated by regulation of genome expression and stability maintenance [
The present issue of BioDiscovery reports an investigation of interphase chromosome architecture in acute myelogenous leukemia by ICS-MCB and its relation to causative translocation between chromosomes 8 and 21 [
Visualizing interphase chromosomes
The interphase chromosome architecture is commonly determined through application of FISH (fluorescence in situ hybridization)-based techniques. As noted below, interphase molecular cytogenetic techniques are usually applied either for analysis of specific genomic loci (using probes for relatively small DNA sequences (rarely >1Mb) comparing to the whole chromosomes) or for painting the whole chromosome, visualized as a chromosome territory (reviewed in [
ICS-MCB is an intriguing alternative to the aforementioned approaches, since it gives an opportunity to determine structure and arrangement of differentially painted chromosomal regions. Its application allows the analysis of chromosome (chromosomal loci) positioning in interphase and chromosomal associations at “subchromosomal” resolution in a given nucleus (for more details see [
Nuclear genome/chromosome organization and disease
Since the introduction of interphase molecular cytogenetics a significant effort has been made to provide comprehensive information about the meaning of nuclear genome (chromosome) organization [
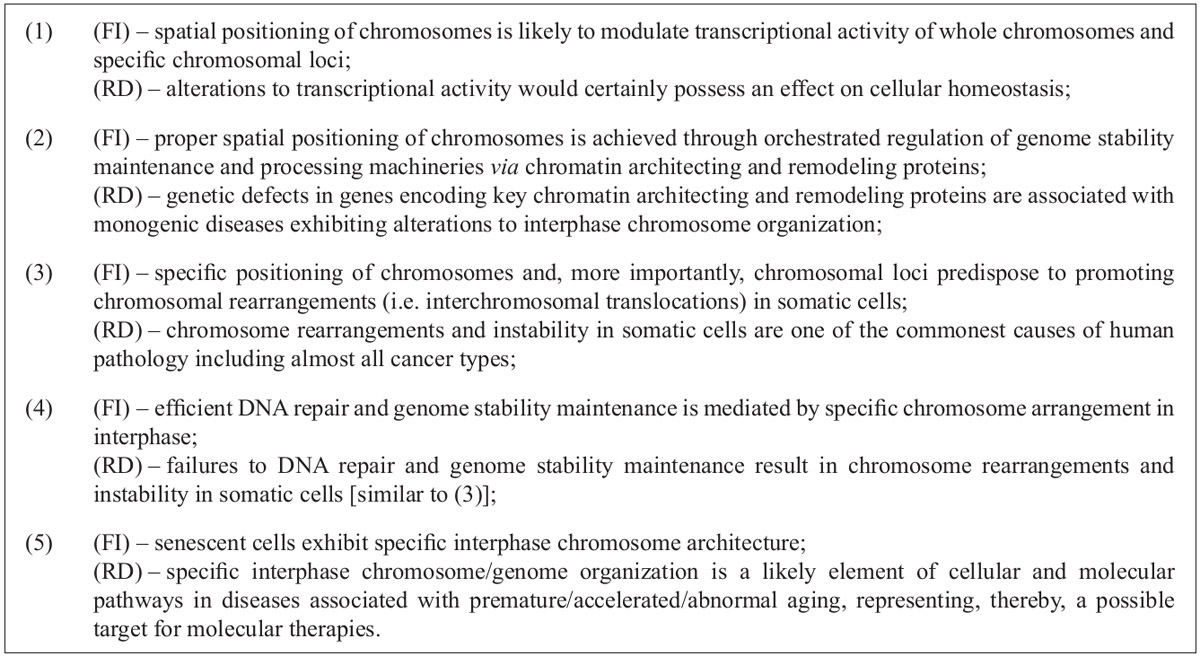
Interphase chromosome architecture plays an important role in modulation of transcriptional activity through chromatin organization [
Although several positive associations between specific interphase chromosome architecture and critical nuclear processes have been made, it is usually hard to come to a definite conclusion concerning pathogenic value of variable chromosome arrangement in interphase nuclei. More probably, specific interphase chromosome organization is rather an element of a pathogenetic pathway rather than a unique underlying disease cause. This idea is further supported by observations on diseases caused by mutations in genes encoding chromatin architecting and remodeling proteins [
Spatiotemporal interphase chromosome organization is highly dynamic [
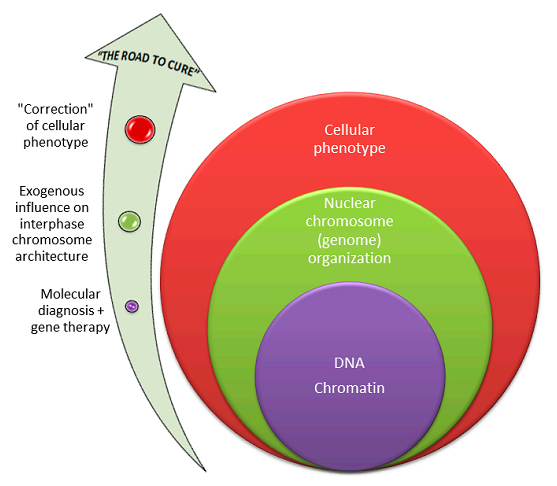
Concluding remarks
Dr. Liehr and colleagues [10] have drawn the attention of BioDiscovery’s readers to the importance of studying interphase chromosome architecture and its associations with pathological conditions. Taking into account the interest to high-order genome organization, positive data seem to be required for encouraging further attempts at characterization of nuclear genome organization in health and disease. Moreover, technological performance of the study, which defines the value of such associations, demonstrates ICS-MCB as the promising method of choice for studying interphase chromosomes at molecular resolution. The anticipated success of related studies is highly dependent on the way how spatial arrangement of interphase chromosomes is determined. Future studies implementing ICS-MCB for evaluation of nuclear genome have the potential to shed light on the role that interphase chromosome architecture does play in disease.
Acknowledgments
I would like to express my gratitude to Professors Svetlana G. Vorsanova and Yuri B. Yurov for their valuable contributions to and helpful discussions during the preparation of this commentary. The author is supported by RFBR grant 12-04-00215-а (Russian Federation, 2012-2014).
References
- Misteli T: Beyond the sequence: cellular organization of genome function. Cell 2007; 128: 787-800.
Reference link - Rajapakse I, Groudine M: On emerging nuclear order. J Cell Biol 2011; 192: 711-721.
Reference link - Vorsanova SG, Yurov YB, Iourov IY: Human interphase chromosomes: a review of available molecular cytogenetic technologies. Mol Cytogenet 2010; 3: 1.
Reference link - Iourov IY, Liehr T, Vorsanova SG, Kolotii AD, Yurov YB: Visualization of interphase chromosomes in postmitotic cells of the human brain by multicolour banding (MCB). Chromosome Res 2006; 14: 223-229.
Reference link - Iourov IY, Liehr T, Vorsanova SG, Yurov YB: Interphase chromosome-specific multicolor banding (ICS-MCB): a new tool for analysis of interphase chromosomes in their integrity. Biomol Eng 2007; 24: 415-417.
Reference link - Manvelyan M, Hunstig F, Mrasek K, Bhatt S, Pellestor F, Weise A et al.: Position of chromosomes 18, 19, 21 and 22 in 3D-preserved interphase nuclei of human and gorilla and white hand gibbon. Mol Cytogenet 2008; 1: 9.
Reference link - Manvelyan M, Hunstig F, Bhatt S, Mrasek K, Pellestor F, Weise A, et al.: Chromosome distribution in human sperm - a 3D multicolor banding-study. Mol Cytogenet 2008; 1: 25.
Reference link - Manvelyan M, Kempf P, Weise A, Mrasek K, Heller A, LierA et al.: Preferred co-localization of chromosome 8 and 21 in myeloid bone marrow cells detected by three dimensional molecularcytogenetics. Int J Mol Med 2009; 24: 335-341.
- Klein E, Manvelyan M, Simonyan I, Hamid AB, Guilherme RS, Liehr T et al.: Centromeric association of small supernumerary marker chromosomes with their sister-chromosomes detected by three dimensional molecular cytogenetics. Mol Cytogenet 2012; 5: 15.
Reference link - Othman M, Lier A, Junker S, Kempf P, Dorka F, Gebhart E et al.: Does positioning of chromosomes 8 and 21 in interphase drive t(8;21) in acute myelogenous leukemia? BioDiscovery 2012; 4: 2.
Reference link - Meaburn KJ, Misteli T, Soutoglou E: Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol 2007; 17: 80-90.
Reference link - Iourov IY, Vorsanova SG, Yurov YB: Chromosomal variation in mammalian neuronal cells: known facts and attractive hypotheses. Int Rev Cytol 2006;249: 143-191.
Reference link - Rouquette J, Cremer C, Cremer T, Fakan S: Functional nuclear architecture studied by microscopy: present and future. Int Rev Cell Mol Biol 2010; 282: 1-90.
Reference link - Strickfaden H, Zunhammer A, van Koningsbruggen S, Köhler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus 2010; 1: 284-297.
Reference link - Schneider R, Grosschedl R: Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 2007; 21: 3027-3043.
Reference link - Ktistaki E, Garefalaki A, Williams A, Andrews SR, Bell DM, Foster KE et al.: CD8 locus nuclear dynamics during thymocyte development. J Immunol 2010; 184: 5686-5695.
Reference link - Misteli T: Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol 2010; 2: a000794.
Reference link - Lever E, Sheer D: The role of nuclear organization in cancer. J Pathol 2010; 220: 114-125.
- Misteli T, Soutoglou E: The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 2009; 10: 243-254.
Reference link - Göndör A, Ohlsson R: Chromosome crosstalk in three dimensions. Nature 2009; 461: 212-217.
Reference link - Bártová E, Jirsová P, Fojtová M, Soucek K, Kozubek S: Chromosomal territory segmentation in apoptotic cells. Cell Mol Life Sci 2003; 60: 979-990.
- Yurov YB, Vorsanova SG, Iourov IY: GIN’n’CIN hypothesis of brain aging: deciphering the role of somatic genetic instabilities and neural aneuploidy during ontogeny. Mol Cytogenet 2009; 2: 23.
Reference link - Mehta IS, Figgitt M, Clements CS, Kill IR, Bridger JM: Alterations to nuclear architecture and genome behavior in senescent cells. Ann N Y Acad Sci 2007; 1100: 250-263.
Reference link - Foster HA, Bridger JM: The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma 2005; 114: 212-229.
Reference link - Mateos-Langerak J, Goetze S, Leonhardt H, Cremer T, van Driel R, Lanctôt C: Nuclear architecture: Is it important for genome function and can we prove it? J Cell Biochem 2007; 102: 1067-1075.
Reference link