Introduction
Embryonic stem cells have attracted great interest both for their use in lab research and possible practical applications [
However, hESCs chromosome preparations frequently encounter certain difficulties, perhaps connected with hESCs biology and chromosome organization, thus making karyotyping problematic [
Materials and methods
Cell lines: Sublines hESM01r18 (46,ХХ,-18,+mar) and hESM0309 (46,ХХ,del(4),dup(9)) were derived from hESM01 and hESM03 parental cell lines, respectively, as described previously [
Metaphase chromosome preparations: Chromosome samples were prepared according to standard procedures. In brief, PBL metaphase and prometaphase chromosomes for FISH analysis were performed as described by Henegariu [
DNA probe preparation: Chromosome microdissection was performed according to previously described methods [
A telomere-specific biotin-11-dUTP labeled (TTAGGG)n DNA probe was generated by PCR as described by Ijdo [
Fluorescence in situ hybridization (FISH): The chromosomal location of the dissected chromosome fragments was assessed by reverse painting as previously described [
Chromosomes were counterstained with 1 µg/ml 4’,6-diamidino-2-phenylindole (DAPI) and analyzed using an AXIOPlan2 Imaging (Carl Zeiss, Jena, Germany), equipped with CCD camera, filter sets, and ISIS5 image-processing package of MetaSystems GmbH. Chromosomes and chromosomal regions were identified by inverted DAPI-banding using human an International Systems for Human Cytogenetic Nomenclature [
Results
Chromosomal rearrangements in hESM0309 cell line
Using DAPI staining, it was shown previously that in all cells of hESM0309 cell line chromosomes 4 and 9 were rearranged [
(i) The breakpoints involved in chromosome 4 deletion formation were characterized as 4q25 and q31.1 (Fig. 1). Based on the data abnormal chromosome 4 was described as del(4)(q25q31.1).
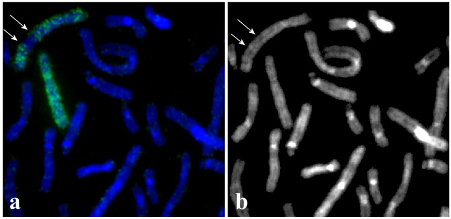
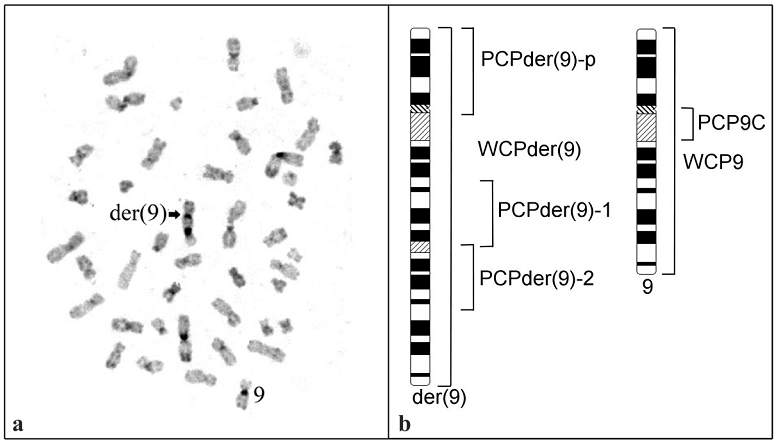
(ii) Another abnormal chromosome found in hESM0309 cells was previously described as dup(9)(pter→q3::q12→qter) by DAPI banding [5]. C-banding revealed two C-positive regions confirming the location of one of the breakpoints in a C-positive region 9q12 (Fig. 2a). To describe chromosome dup(9) more precisely the set of DNA probes were generated by microdissection from this chromosome and normal chromosome 9 as shown schematically in Fig. 2b. E.g. DNA probes WCP9 and WCPder(9) (Fig. 2b) completely painted chromosome 9 and dup(9), however, the intensity of the FISH staining of these chromosomes was different (Fig. 3). Further studies summarized in Fig. 3 revealed a dup(9)(pter→q33::q12→qter) in hESM0309.
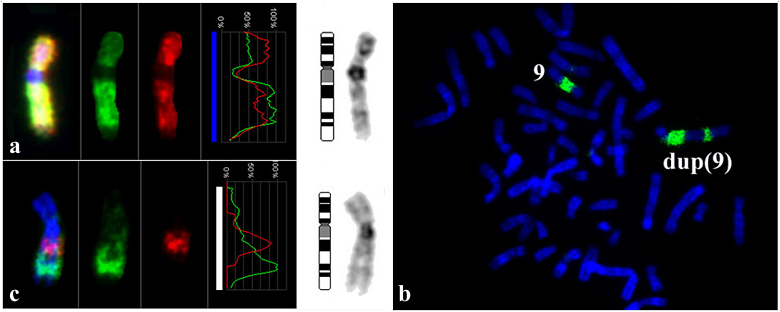
Thus, the karyotype of hESM0309 cells was described 46,XX,del(4)(q25q31.1),dup(9)(q12q33).
Chromosomal rearrangements in hESM01r18 cells
A previously performed hESM01r18 karyotype analysis allowed to hypothesize that the marker, most likely an abnormal ring chromosome was a derivative of normal chromosome 18 and hESM01r18 cells were described as 46,ХХ,-18,+?r(18) [
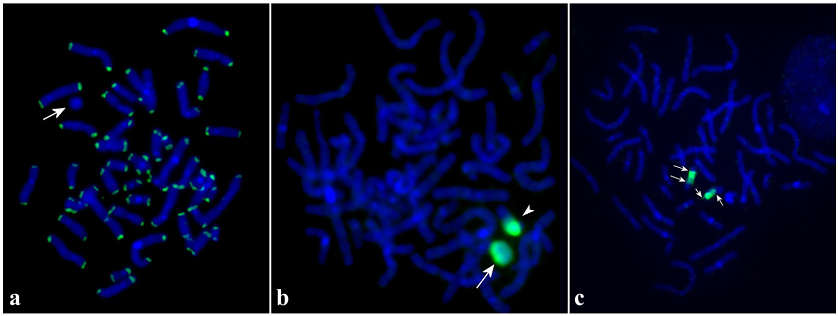
Thus r(18) contain two copies of the indicated region and the karyotype of hESM01r18 cells was 46,ХХ,r(18)(::p11.31→q21.2::q21.2→p11.31::).
Discussion
Since their isolation in 1998 [
Karyotyping of hESM01r18 and hESM0309 cell lines demonstrated that chromosomes r(18) and dup(9), respectively, were present in all cells from given cell lines [
The breakpoints in chromosome 9 revealed in hESM0309 cell line coincides with the breakpoints quite often observed during loss of heterozygosity in transitional cell carcinoma of urinary tract tumor (9q12, 9q22.3, 9q33-34) [
The existence of chromosomal abnormalities in the stem cells are often associated with carcinogenesis [
Overall, this study shows that hESCs should be (i) molecular cytogenetically characterized in detail and (ii) that such studies may be extremely helpful in understanding tumor initiation and progression, as well.
Acknowledgments
This work was supported by Russian Federal contract 02.512.11.2060.
References
- Corrales NL, Mrasek K, Voigt M, Liehr T, Kosyakova N. Copy number variations (CNVs) in human pluripotent cell-derived neuroprogenitors. Gene 2012; 506(2): 377-379.
Reference Link - Maitra А, Arking D, Shivapurkar N, Ikeda M, Stastny V, Kassauei K et al. Genomic alterations in cultured human embryonic stem cells. Nature Genetics 2005; 37: 1099-1103.
Reference Link - Mitalipova М, Rao R, Hoyer D, Johnson J, Meisner L, Jones K. et al. Preserving the genetic integrity of HESCs. Nature Biotechnology 2005; 23(1): 19-20.
Reference Link - Prokhorovich MA, Shilov AG, Rubtsov NB, Kiselev SL, Lagar’kova MA. Rapid and efficient method karyotyping human embryonic stem cells. Cell culture. Newsletter. St. Petersburg 2006; 21: 55–58.
- Prokhorovich MA, Lagarkova MA, Shilov AG, Karamysheva TV, Kiselev SL, Rubtsov NB. Cultures of hESM human embryonic stem cells: chromosomal aberrations and karyotype stability. Bull Exp Biol Med 2007; 144(1): 126-129.
Reference Link - Rooney D.E., Czepulkowski B.H. Human cytogenetics. A practical approach. Oxford Univ. Press 1992;1: 157-192.
- Henegariu O, Heerema N, Wright l, Bray-Ward P, Ward D, Vance G. et al. Improvements in cytogenetic slide preparation: controlled chromosome spreading, chemical aging and gradual denaturing. Cytometry 2001; 43: 101-109.
- Rubtsov N, Senger G, Kuzcera H, Neumann A, Kelbova C, Junker K, Beensen V, Claussen U Interstitial deletion of chromosome 6q: precise definition of the breakpoints by microdissection, DNA amplification, and reverse painting. Hum Genet 1996; 97(6): 705-709.
Reference Link - Karamysheva TV, Matveeva VG, Shorina AP, Rubtsov NB (2001) Clinical and molecular cytogenetic analysis of a rare case of mosaicism for partial monosomy 3p and partial trisomy 10q in human. Genetika 2001; 37: 811-816.
- Telenius H, Carter N, Bebb C Degenerate oligonucleotide-primed PCR: general amplification of target DNA by single degenerate primer. Genomics 1992; 13: 718-725.
Reference Link - Rubtsov N, Karamysheva T, Babochkina T, Zhdanova N, Trifonov V, Starke H. et al A new simple version of chromosome microdissection tested by probe generation for 24-multi-color FISH, Multi-color banding (MCB), ZOO-FISH and in clinical diagnostics. Medgen 2000; 12: 65.
- Ijdo J, Wells R, Baldini A and Reeders S. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Research 1991; 19: 4780.
Reference Link - Lichter P, Cremer T, Borden J, Manuelidis L, Ward D. Delineation of individual human chromosomes in metaphase and interphase cells by in situ supression hybridization using recombinant DNA libraries. Hum Genet 1988; 80: 224-234.
Reference Link - An Internatinal Systems for Human Cytogenetic Nomenclature (ISCN 2009) Ed. Mitelman F. Basel. S. Karger 2009; 114.
- Thomson J, Itskovitz-eldor J, Shapiro S, Waknitz M, Swiergiel J, Marshall V. et al Embryonic stem cell lines derived from human blastocysts. Science 1998; 282: 1145-1147.
Reference Link - Kimura F, Florl A, Seifert H, Louhelainen J, Maas S, Knowles M, Schulz W. Destabilization of chromosome 9 in transitional cell carcinoma of the urinary bladder. Br J Cancer 2001; 85: 1887-1893.
Reference Link - Murga Penas EM, Callet-Bauchu E, Hongtao Ye, Hinz K, Albert N, Copie-Bergman C. et al. The translocations t(6;18;11)(q24;q21;q21) and t(11;14;18)(q21;q32;q21) lead to a fusion of the API2 and MALT1 genes and occur in MALT lymphomas. Haematologica 2007; 92(3):405-409.
- D’Achille P, Seymour JF, Campbell LJ. Translocation (14;18)(q32;q21) in acute lymphoblastic leukemia: A study of 12 cases and review of the literature. Cancer Genet Cytogenet 2006; 171: 52-56.
Reference Link - Gebhart E, Liehr T Patterns of genomic imbalances in human solid tumors (Review). Int J Oncol 2000; 16: 383-99.