1. Introduction
1.1. Basic definitions
Warms up and finally burns.
Thoughts of Vantala (c. ХІІ century).
Unlike all other polymers in living cells, DNA cannot be completely broken down to monomers and rebuilt from scratch. Its synthesis always requires a pre-existing single strand of DNA as a template and a double-strand region at least several bp long, providing a 3'-OH group from which the synthesis to start. The DNA of the cell is the single blueprint that contains all the information on how to make a new copy of any other polymer molecule in the cell. Therefore, it is sensible to make sure that the genetic information coded in DNA could always be recovered even from a single strand. The preservation of the genetic information in its original state is dependent not only on the accuracy of its copying during DNA replication, but also on the timely repair of any event which may compromise the integrity of the DNA molecule and/or may change its building blocks or their arrangement in any manner. Such events constitute DNA damage.
The definitions of DNA damage are numerous in the literature. One of the most succinct but, at the same time, complete definitions is the definition given by Benjamin Lewin in his famous 'Genes' textbook:
"Damage to DNA consists of any change that introduces a deviation from the usual double-helical structure" [
One of the most commonly used and, in the authors' opinion, the most comprehensive, is the definition of DNA damage given by the US National Library of Medicine, [Unified Medical Language system (UMLS)]:
"Drug- or radiation-induced injuries in DNA that introduce deviations from its normal double-helical conformation. These changes include structural distortions which interfere with replication and transcription, as well as point mutations which disrupt base pairs and exert damaging effects on future generations through changes in DNA sequence".
The latter definition, however, implies that DNA damage is always a product of some external influence, that is, it is not produced by physiological means. This is not true, however, as the most part of DNA damage occurs as a result of normal cellular metabolism. Therefore, for the purposes of the present book, we may adopt the following definition of DNA damage:
"Any modification in the physical and/or chemical structure of DNA resulting in an altered DNA molecule which is different from the original DNA molecule with regard to its physical, chemical and/or structural properties".
To understand more fully the mechanisms of repair of DNA, one needs to be familiar with the basic types of DNA damage and, respectively, the agents which cause it. Basic types of DNA damaging (genotoxic) agents classified by their essential properties are presented below.
1.2. Classification of factors causing DNA damage
1.2.1. Factors of exogenous and endogenous origin
DNA damage may occur under the influence of factors external to the cell (factors of exogenous origin, e.g. environmental factors) or potentially aggressive agents produced by normal cell metabolism (factors of endogenous origin). The consequences for the cell's DNA damage caused by the action of endogenous factors may be more serious and/or more extensive than the effect of most of the exogenous DNA damaging factors. DNA damaging events caused by endogenous factors generally occur much more frequently than damage caused by exogenous factors. For example, several thousands of nitrogenous bases are lost daily from DNA in eukaryotic cells as a result of spontaneous base hydrolysis alone.
DNA damaging factors of endogenous and of exogenous origin may exert their genotoxic action on DNA using the same mechanism. For example, both ionising radiation (an agent of environmental origin) and normal oxidative phosphorylation (obviously of endogenous origin) generate reactive oxygen species which may damage the cell's DNA.
DNA damage may cause appearance of structures that do not occur naturally in DNA or may cause structures that are perfectly normal for some DNA regions, to appear at ectopic sites. For example, thymine dimers are structures which normally do not exist at all in undamaged DNA. Occurrence of triple-strand DNA regions is perfectly in order in certain genome areas, but may accidentally arise de novo in other areas, where it constitutes damage.
DNA damaging agents may work directly on DNA (e.g. base modifying agents) or may induce structural changes that increase the risk for further damage. For example, the occurrence of triple-strand structures in DNA increases the risk for copying errors during subsequent replication of the affected DNA region (see below).
1.2.2. Nature of DNA damaging agents (physical and chemical agents)
Genotoxic agents may be broadly classified into factors of chemical or of physical nature, respectively. Some genotoxic agents are capable of damaging DNA because of both their chemical and their physical properties. The damaging agents acting on DNA by virtue of their physical properties are, for example, short-wavelength electromagnetic energy such as ultraviolet (UV) radiation and ionising radiation. Under specific conditions, low-energy electromagnetic waves, such as infrared radiation (heat) and microwave and radio wave radiation may also cause DNA damage [
DNA damaging agents may work directly on DNA to alter its structure or may exert its activity via inducing structural change which, in turn, alters its properties, or these two may occur simultaneously. For example, agents introducing bulky adducts in DNA work both ways, as they may chemically modify the DNA molecule and at the same time may physically alter (bend, distort, twist) its structure. The latter may increase the risk for additional damage (e.g. introduction of strand breaks).
1.2.3. Factors causing alterations in the information content of DNA and factors causing structural alterations
Damage in DNA is often, but by no means always accompanied by alteration of the information content of DNA. Only certain damaging events, left unrepaired, can produce a lasting, heritable change in the DNA sequence, thus producing a mutation. Among these are, for example, events producing a mismatch in DNA. Mismatched bases are usually recognised and promptly repaired by a designated mismatch repair system, but if the DNA template containing the mismatches is replicated despite the presence of damage, the resulting daughter strands will carry different 'meaning' in their DNA. Since there is no system in the cells that could recognise which daughter molecule is identical to the 'maternal' molecule and which is different, the cell carrying the alteration would faithfully copy the altered sequence during subsequent cell divisions.
DNA damage or erroneous copying of the damaged fragment may change one or more of the building blocks of DNA, but the resulting altered variant may carry the same coding sense. Some types of DNA damage, if they are left unrepaired, would cause replication blockage. Generally, 'nominal' DNA polymerases of replication would stop when they encounter damage in the template. This would, in turn, result in recruitment of the repair machinery to the blockage site, and the damage would be repaired without any lasting changes in the DNA sequence. Other types of DNA damage (for example, some of the events producing modified nucleotides) are sometimes managed in a manner that accommodates the modified nucleotide/s - that is, they are not exactly ignored, but during replication the fragment containing the modified nucleotide/s is copied in a manner that restores the original sequence, despite the fact that the template is damaged. DNA damage may affect structures of higher order than the nucleotide sequence level. For example, the occurrence of regions with Z-conformation or the already mentioned triple-strand DNA may constitute DNA damage. DNA damage involving higher-order structures may affect crucial cellular processes, but may not be transmitted to the cellular progeny during replication, as the nucleotide sequence remains essentially the same. Other types of alteration of higher-order structures may result in inaccurate copying during replication, which may produce a heritable genetic change (that is, a mutation).
1.2.4. Cytotoxic and genotoxic agents
Factors of any origin which are injurious to living cells may be damaging to the cell as a whole (cytotoxic agents) or specifically damaging to the cellular DNA (genotoxic agents). The same agent may have predominantly cytotoxic or predominantly genotoxic action in living cells depending on the type of the agent, the type of the cells in question and the duration of the action of the damaging agent. For example, ionising radiation may be cytotoxic in large doses, that is, cells and cell populations may die directly as a result of the massive oxidative stress caused by rapid elevation of the levels of free radical species in the cell. In smaller doses, however, ionising radiation may exhibit only genotoxic properties - it produces significant amounts of DNA damage, but not all damaged cells would die, neither straight away nor as a late effect. Same is the case with many chemical damaging agents. For example, aromatic amines (e.g. 2-acetylaminofluorene) in lower concentrations exhibit predominantly genotoxic properties, as they work by introducing bulky adducts in DNA, which makes copying of the templates carrying those adducts difficult and more error-prone than copying undamaged DNA. In larger doses, however, aromatic amines may be directly cytotoxic [
Finally, cytotoxic effects may be exerted via genotoxic action. For example, most chemotherapeutic anticancer agents work by introduction of significant amount of damage in DNA in the hope that this may cause rerouting of the cancer cells towards the programmed cell death pathway.
1.2.5. Mutagens and teratogens
General George S. Patton (1885-1945)
Those two terms are often used synonymously or, at least, interchangeably, but this is incorrect most of the times. There is an essential difference between the intended targets and sometimes in the modes of action of mutagenic and teratogenic agents. Mutagen is a term that denotes an agent that directly or indirectly affects DNA so that may eventually cause a potentially heritable change in its sequence or structure. Mutagens usually have inherent carcinogenic properties, as the damage caused by their action may trigger neoplastic transformation. Teratogens are by definition agents that may induce abnormal prenatal development or may increase the risk for such. This, however, may or may not be related to any change or damage in DNA. For example, thalidomide is known to be a powerful teratogen (as, unfortunately, was found out the hard way in the 1950's) but is not a mutagen, as it works by interference with the vascularisation of developing body parts during the morphogenesis of the embryo. There is no DNA damage involved whatsoever. UV light is powerful somatic mutagen but is usually not considered to be a teratogen, as embryos inside the womb are very well protected from UV irradiation. It is understood, however, that the same agent may exhibit both mutagenic and teratogenic properties - for example, ionising radiation or chemical agents such as formaldehyde may very well cause both somatic mutagenesis and teratogenic effects, but the one does not automatically include the other as well. The fluorescent dye ethidium bromide, commonly used in molecular biology laboratories, is considered to be a powerful somatic mutagen (intercalating agent) but its teratogenic properties have not been definitely proven. It is possible that the usual mode and level of exposure (usually, skin coming into occasional contact with diluted solutions) minimise the potential teratogenic effects. Ethidium bromide was regularly administered in the past in veterinary medicine as an antiprotozoal drug (specifically, against Trypanosoma spp.), and had been used for over 50 years before being replaced by more modern drugs [
There are special circumstances, however, in which an agent that would normally demonstrate only mutagenic or only teratogenic properties may occasionally exhibit the other type of properties. This usually involves a known somatic mutagen acting as a teratogen under certain conditions. This is usually related to dose- or stage-sensitivity phenomena. For example, the already mentioned UV light, a known somatic mutagen, has been documented to actually have weak teratogenic properties. Specifically, it has been associated with a slight increase in the risk for neural tube defects for children of parents who have been exposed to therapeutic doses of UV (specifically, during phototherapy for psoriasis). The teratogenic effects are reportedly exerted via modulation of the folate metabolism and are usually restricted to very early pregnancy (the first weeks post-conception) [
When tested for mutagenic properties (e.g. using the classic Ames test [
1.3. DNA damage does not always result in mutagenesis
when a thing hits, but not when it misses.
Francis Bacon Sr., (1561-1626)
Undoubtedly, an event that occurs tens of thousands times per day can be described as 'common'. Therefore, DNA damage is a very common event indeed. The bulk of this damage is repaired promptly and accurately by the cellular repair machinery. This machinery, however, is not perfect (becoming even less so with ageing), and it may occasionally miss sites of damage or make mistakes - for example, it may add a non-matching nucleotide.
Most mismatches in DNA result from errors in template copying during DNA replication. As DNA repair is almost always associated with DNA synthesis, it can produce mismatches as well. The error rate in repair-related DNA synthesis is generally higher (≈10-3-10-6 per nucleotide per cycle of synthesis for different 'repair' DNA polymerases and for different types of mispairs) than the error rate in replication (10-5-10-7 per nucleotide per cycle of replication) [
Unrepaired damage in DNA may eventually bring about the death of the cell (if the damage is too extensive and/or too severe) or it may be retained, tolerated and, if the cell is capable of division, potentially transmitted to the cell's progeny. There are several DNA polymerases capable of copying damaged templates, albeit with a higher error rate, collectively known as the Y-family of polymerases. In prokaryotes, these are DNA polymerases IV and V [
In the process of copying a damaged template, the DNA polymerases may add the correct nucleotide and synthesise a complementary strand which is identical in sequence with the template strand; or they may add a mismatched nucleotide, synthesising a daughter strand which carries altered nucleotide sequence at the site where the damage initially occurred. During the next cycle of replication, the altered DNA molecule will produce daughter molecules carrying the altered sequence, transmitting it on to their progeny. Eventually, a mutant clone is created. A genetic alteration which is capable of being transmitted down the generations is called a mutation. Therefore, it is prudent to assume that the main difference between DNA damage and DNA mutation is that DNA mutations are heritable, while DNA damage may be not.
Generally, less than 0.1% (1 out of 1000) of the base substitutions occurring in DNA are retained and, potentially, transmitted to cell progeny. That means that over 99.9% of the damage occurrences are detected and repaired so that no trace of them is left whatsoever. The rate of spontaneous mutation varies between different organisms, but the average estimates may be summarised as follows:
- Viruses with RNA genomes (genome length ≈104 nucleotides) - about 10-4-10-5 per viral genome per cycle of replication;
- Viruses with DNA genomes - for example T4 phage (double-stranded DNA genome, 3x105 bp in length) - about 10-6-10-8 per viral genome per cycle of replication;
- Bacteria (for example E. coli, genome length ≈106 bp) - about 10-5-10-10 per cell cycle;
- Monocotyledon plants (for example maize, haploid genome about 2x109 bp in length) - about 10-6-10-7 per base per generation;
- Invertebrates (for example the fruit fly D. melanogaster, haploid genome about 1.4x108 bp in length) - about 10-7-10-8 per base per generation;
- Mammals, including humans (nuclear haploid genome about 3x109 bp in length) - about 10-8 per base per generation.
Mitochondrial DNA (1.66x104 bp in humans) has a mutation rate of its own, which is higher than the mutation rate in nuclear DNA - about 10-5-10-6 per base per generation [
It is easy to calculate that since the incidence of spontaneous mutations in higher eukaryotes is about 10-8 per base per generation, in an average genome (about 109 bp), every cell division would result in about a dozen mutations [
Spontaneous mutagenesis is generally a random process. Even among non-synonymous mutations, resulting in substitutions of amino acid residues in the resultant protein, however, there might be a mutation bias - that is, some nucleotides and some codons may be more frequently affected by mutations than others. For example, it was demonstrated that 30% of the mutations resulting in genetic disease in man affect specifically arginyl and glycyl residues [
It has been proposed that the two major factors for cellular ageing are the accumulation of mutations in DNA resulting from replication errors (nucleotide substitutions, nonsense mutations); and the oxidative stress resulting from the genotoxic action of reactive oxygen species (ROS) on mitochondrial DNA [
1.4. Accumulation of mutations in DNA may be responsible for 'diseases of old age'
Maurice Chevalier (1888-1972)
Despite the fact that the rate of occurrence of DNA damage is very high, the overall mutation rate is very low, thanks to the efficiency of the mechanisms for recognition and repair of damage in normal cells. Every cell cycle is associated with the production of new mutations in the cell's DNA, but discernible signs of ageing or age-related disease (degenerative diseases or cancer) appear after some time, that is, after the process has been repeated a sufficient number of times so that enough potentially harmful mutations have accumulated.
It is currently believed that accumulation of mutations in eukaryotic DNA may account for the increased prevalence of cancer at advanced age. It was already mentioned, however, that beyond certain age the cancer rate in man actually decreases, and the prevalent causes of mortality in the oldest old are other diseases and conditions, such as infections or multiple organ failure (the latter being the essence of 'dying of old age'). One can suppose that individuals who have lived to >85 years of age without developing cancer of any type must possess a very efficient system of DNA repair which allows them to deal with DNA damage in timely manner so that potentially carcinogenic mutations accumulate at a slower rate.
'Dying of old age' has a lot to do with the capacity to repair DNA damage, but in a different way. Apart from damage repair, other mechanisms for dealing with DNA damage exist in eukaryotic multicellular organisms. These mechanisms are activated when the damage is too extensive or too severe to be repaired. They generally work by inducing permanent cell cycle arrest (replicative senescence) and/or by elimination of the damaged cell from the pool (programmed cell death/apoptosis). Apoptosis is usually completed quickly, sometimes within hours. A cell that has been rendered incapable of further division may live and function for a long time, sometimes weeks, months and years. Eventually, however, it would die. Regardless of the exact mechanism by which a cell died, however, it must be replaced, so that the tissue would continue to function normally. Upon the loss of damaged somatic cell/s, the resident stem cells of the tissue (adult stem cells) are stimulated to divide, producing new precursor cells that would take the pathway of differentiation, eventually becoming specialised cells (or, more typically, producing precursor cells that would undergo multiple divisions during their differentiation, producing many specialised cells). Stem cells in adult tissues, however, have a limited capacity for division, that is, like all somatic cells, their ability to produce new precursor cells diminishes with age and mutations accumulate with every cell cycle. True stem cells in adult tissues are supposed to last for a lifetime; therefore, they are induced to divide only infrequently. The capacity for multiple divisions is usually delegated to their direct descendants - the precursor cells. As age advances, the capacity of adult stem cells to repopulate the tissue they belong to slowly declines, eventually reaching a threshold beyond which the tissue or the organ cannot implement its functions normally. Degenerative disease develops, culminating in organ failure that would eventually kill the multicellular organism. The prevalence of degenerative disease increases as age advances. Efficient DNA repair may stave off the onset of age-related disease, but not indefinitely. It has been proposed that cellular senescence (and its larger-scale counterpart, ageing) have been specifically developed by Nature to ensure that multicellular living creatures are kept healthy during the phase of life in which they are most likely to contribute to the genetic pool; and then removed in order to let their offspring take their place.
1.5. A la guerre comme à la guerre, or how well does DNA repair perform under pressure
Albert Pike (1809-1891)
DNA repair works very efficiently in managing everyday DNA damage. How efficient is damage repair, however, in conditions of a severe genotoxic attack? The answer is, again, "very efficient", though within certain limits and dependent on the severity and the duration of the genotoxic effects, the cell type, and the species in question. For example, Deinococcus radiodurans can withstand over ten thousand of Gy with only about 40-50% reduction in survival rates [
Extreme radioresistance is not limited solely to bacteria. It is widely known that some invertebrates, such as Tardigrade may resist up to 5000Gy of ionising radiation [
The resistance of different cell types to genotoxic attacks may greatly vary. Generally, the higher is the proliferative potential of a cell, the higher its sensitivity to genotoxic agents. Cell that divide rarely (or practically never), such as differentiated neurons, are generally more radioresistant than cells with naturally rapid turnover (blood cells, epithelial cells). Differential resistance to DNA damaging agents is in the basis of modern anticancer therapy.
There have been case reports of higher animals and even humans demonstrating remarkable resistance to genotoxic treatments - to an extent, of course. For example, large doses of UV may cause short-term as well as long-term consequences in man, starting from erythema and desquamation and ending with severe eye disease and skin cancer. It is unlikely, however, that UV irradiation would be immediately life-threatening. During a course of cytostatic therapy, an individual may ingest several lethal doses of a genotoxic drug, exhibiting only some adverse effects. As for ionising radiation, though exposure is always undesirable, there are remarkable examples for radioresistance as well. For humans, a whole-body acute dose of 5Gy is in most cases lethal. Humans, however, can survive doses of 10-15Gy when administered in fractionated doses of 1.5-2Gy - with proper medical care, of course. Such dosages are often prescribed in myeloablative regimens before transplantation of haematopoietic stem cells for treatment of haematological cancers.
Apart from iatrogenic genotoxicity, there are more notable cases of human radioresistance, though they are usually a product of an accident. For example, among the people who were on site when the Chernobyl incident happened and who were directly involved in the immediate crisis management (e.g. fire-fighters), several survived after receiving acute doses of 4-5Gy [
2. Types of DNA damage
And safety is among them.
Johann Wolfgang von Goethe (1749-1832)
Damage to DNA may occur as a result of normal cellular metabolism (endogenous damage) or under influences of origin external to the cell (exogenous damage). Therefore, DNA damaging agents and the associated damage mechanisms may be broadly classified as agents/mechanisms of endogenous or of exogenous origin.
2.1. Damage of endogenous origin
2.1.1. Conversion of one base to another, producing a mismatch
The four nitrogenous bases in DNA may be subject to direct conversion into one another or into rare bases which may have pairing affinity to bases different from the original pairing partner (non-canonical pairing). Base conversion often results from hydrolysis of nitrogenous bases in DNA.
Deamination of nitrogenous bases is a very common type of hydrolytic damage in DNA. For example, deamination of cytosine produces uracil (Fig. 1).
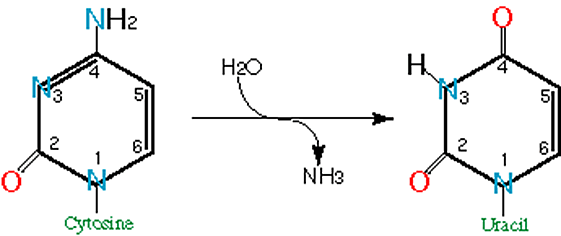
Uracil pairs more efficiently with A than with G, thereby creating a mismatch. In the next round of DNA replication, this will result in a substitution of a C:G pair with an A:T pair.
Adenine may be spontaneously deaminated to hypoxanthine (Fig. 2), the latter pairing more readily with C than with T.
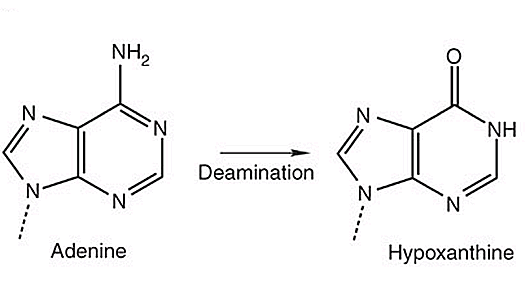
Similarly, guanine deaminates to xanthine, which pairs to thymine instead of cytosine. In both cases deamination results in substitutions of C:G pairs to A:T pairs or vice versa.
Modified bases may mispair with partners different from their 'canonical' partners. For example, 5-hydroxyuracil, product of oxidative deamination of cytosine, would pair with G, but also will mispair with any with the other three nucleotides, albeit with somewhat lower affinity [
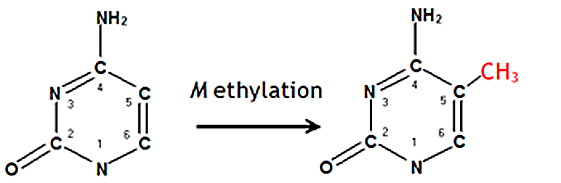
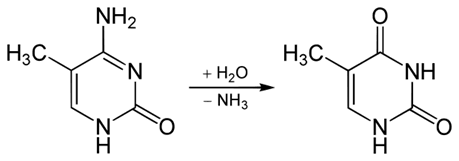
Base conversions may also result from modification of bases which have already been affected by some other type of modification. For instance, methylation of cytosine on position 5 in the heterocyclic ring produces 5-methylcytosine (Fig. 3a). The resulting 5-methylcytosine may be subsequently deaminated, producing thymine (Fig. 3b) which pairs in a canonical manner with adenine, substituting a C:G pair with a T:A pair in the next round of replication.
Deamination of 5-methylcytosine is a fairly common event, occurring with a frequency no lower than 102 events per mammalian cell per day [
Analogues of normal nitrogenous bases may also pair with partners other than the 'canonical' partners. For example, both adenine, which is a 6-aminopurine, and its analogue 2-aminopurine pair with T, but 2-aminopurine may also occasionally pair with C instead.
Mismatched bases in DNA are among the critically important alterations of the information content of DNA. Substitution of bases may produce substitutions of amino acids; therefore, alter partially or completely the properties of the resulting protein. Alternatively, mismatches may introduce a premature stop signal in mRNA, resulting in production of truncated and/or unstable protein or no protein at all. There are designated mechanisms for repair of mismatched bases on DNA. Regions with high CG content (CpG islands) are found in the promoter regions of about 40% of mammalian genes and are generally associated with the control regions of genes [reviewed in
2.1.2. Loss of nitrogenous bases due to hydrolysis
Typical example of hydrolytic loss of nitrogenous bases is DNA depurination, a very common type of DNA damage occurring spontaneously 5x103 - 1x104 times per genome per day in human cells [
Loss of nitrogenous bases in DNA is strongly dependent on the temperature. At t=37°C, the E. coli genome (length in the order of 106 bp) loses on the average one base per cell generation, while at t=85°C the genome of Thermus thermophilus (which is, like E. coli's genome, about 106 bp in length) may lose up to 300 bases per cell generation [
Loss of nitrogenous bases results in abasic sites in DNA. This, in turn, may promote strand breakage and/or mispairing.
2.1.3. Alkylation (most commonly, methylation) of nitrogenous bases
Base alkylation usually affects a nitrogen or an oxygen atom in a nitrogenous base in DNA, producing a variety of modified bases - 5-methylcytosine, 7-methylguanine, 1-methyladenine, etc. as well as alkyl phosphates. Alkylated bases may have different pairing properties from their unmodified counterparts. For example, O6-alkylguanine and O4-alkylthymine tend to pair with non-canonical partners (T and G, respectively), producing a substitution of a G:C to an A:T pair and vice versa [
The alkyl modification is usually in the form of methylation on selected positions in the nitrogenous bases in DNA. While methylation of a base in DNA constitutes a minor structural alteration by itself, the outcomes of its presence in DNA may be very serious. As it was already mentioned, the modified base may be subjected to further modification, efficiently converting one base to another which may later result in DNA mismatch (see below). Even if the presence of the alkylated base in DNA does not increase the risk for base substitutions, it still may produce long-term effects without any further chemical modification. For example, methylation at selected nucleotides may trigger alterations of the chromatin architecture around the site where the modification occurred. This may have many and varied consequences. Methylation at cytosine residues in CpG islands may result in inactivation of the expression of the gene/s under the control of the respective CpG island, as methylation on specific residues in the control regions of genes is a routine mechanism for switching gene expression off. Deregulated DNA methylation is a common finding in tumours. It is believed that it is a microevolutionary event in cancer, as some genes (e.g. coding for products suppressing cell proliferation or acting in DNA repair, such as cyclin kinase inhibitors, BRCA1, retinol-binding protein-1, angiogenesis inhibitor, O6-methylguanine methyltransferase, etc.) must be switched off while others (e.g. genes coding for products which act as positive regulators in cell division, such as growth factor receptors) must be constantly expressed in order to ensure the growth of the tumour [
Disorders of methylation may affect not only somatic cells (as in cancer), but also the germline. Several inherited human diseases have been shown to be associated with aberrant methylation. Among these prominent is Rett syndrome, a rare neurodevelopmental disorder caused by mutations in the gene encoding methyl-CpG-binding protein-2 (MECP2), a protein that recognises methylated bases in DNA [
2.1.4. Oxidation of nitrogenous bases
Oxidation of nitrogenous bases affects the purine bases (A, G) as well as the pyrimidines (C, T). Purine base oxidation results in 8-oxopurines (Fig. 4, upper row on the left) or, alternatively, if the imidazole ring is opened, in formamidopyrimidines (Fig. 4, lower row on the right). Oxidation products of pyrimidines are usually thymine glycol, uracil glycol, 5-hydroxycytosine and 5-hydroxyuracil (Fig. 4, upper row on the right and middle row) [
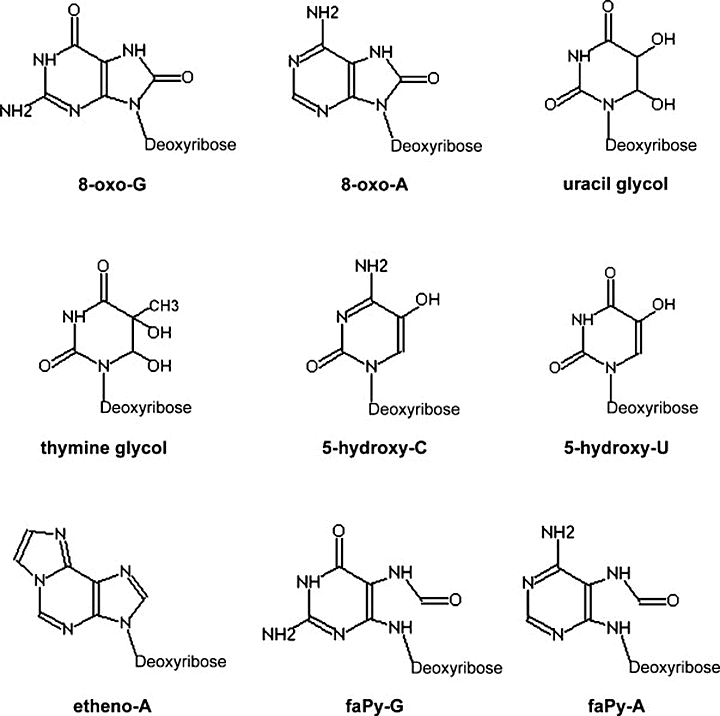
8-oxo-G : 7,8-dihydro-8-oxo-guanine
8-oxo-A : 7,8-dihydro-8-oxo-adenine
5-hydroxy-C: 5-hydroxycytosine
5-hydroxy-U: 5-hydroxyuracil
etheno-A: ethenoadenine
faPy-G: 6-diamino-4-hydroxy-5-formamidopyrimidine
faPy-A: 4,6-diamino-5-formamidopyrimidine [
Base oxidation in DNA is often accompanied by strand breaks (see below), as the same damaging agent (most often, free radical species) can cause both types of DNA damage.
Accumulation of oxidative DNA damage (oxidative stress) is believed to be one of the major mechanisms in ageing and disease.
2.1.5. DNA breaks of endogenous origin
Single-strand breaks (SSBs) result from disruption of the phosphodiester bond between two adjacent deoxyribose residues in the backbone of DNA. SSBs are among the most common instances of DNA damage. Double-strand breaks (DSBs) are less common in living cells, and their presence is generally not tolerated.
Strand breaks frequently occur as a result of normal manipulation of DNA during nuclear transactions (untangling DNA during transcription and replication, relaxation of supercoils, etc.). These are, however, usually repaired as part of the process of DNA manipulation, as the enzymes which catalyse the strand breakage (topoisomerases) possess an intrinsic ligation activity as well, allowing them to rejoin the broken strands [
Breaks in DNA and specifically DSBs constitute a potent signal for recruitment of the DNA repair machinery at the break site. DNA breaks are normally promptly repaired, unless the mechanisms for recognition or processing of damage of this type are faulty (as is in some inherited diseases associated with genomic instability). Presence of DSBs may be normal for some types of cells, but it is generally restricted to a specific phase of the life cycle of the cell. For example, DSB occurrence is normal in germ cells during meiosis, as it ensures exchange of DNA segments between non-sister chromatids of homologous chromosomes. Also, generation of double-strand breaks is a normal part of the process of development of innate immunity in mammals. Specifically, the differentiation of T-cells and B-cells of the immune system requires introduction of multiple DSBs in their DNA during the process of genomic rearrangements requisite to immunoglobulin class switch recombination (V(D)J recombination).
Generally, the clinical presentation of human diseases and conditions associated with deficiencies of repair of DNA breaks virtually always includes some type of immune disorder and/or predisposition to cancer. Some types of severe combined immune deficiency (SCID) occur because of failed V(D)J recombination during the rearrangement of the T-cell receptor genes in T cells and the immunoglobulin genes in B cells [
2.2. Damage of exogenous origin
Exogenous agents may cause many different types of damage to DNA, depending on the nature of agent and the substrate on which it works. Some of this damage is specific to the action of exogenous agents only (e.g. dimerisation) while others may be caused by endogenous factors as well (e.g. base alkylation, strand breaks, etc.)
2.2.1. Dimerisation
Dimers of any type do not normally exist in DNA. Virtually all types of electromagnetic radiation (EMR) may cause dimerisation between bases in DNA, though dimer formation usually results from electromagnetic radiation in the UV range. EMR with energy higher then UV such as ionising radiation (IR) preferentially produces DNA breaks (directly as well as indirectly), whereas lower-energy IR usually exerts its genotoxic effects on DNA by increasing the amount of free radical species in the cell (oxidative damage).
The type of UV-induced damage to DNA is dependent on the wavelength of the UV. Dimers are most often caused by high-energy, short-wavelength UV in the range of 100-300nm (UV-B). Under certain conditions, UV-A light (lower energy, longer wavelength than UV-B (300-400nm)) may also cause direct damage to DNA, including dimers, although the risk for this type of damage is lower than with UV-B. Therefore, when choosing sunscreen products it is sensible to prefer those with UV-A seal or labelled as 'broad-spectrum' (which means that UV-A is screened out as much efficiently as UV-B), as there is a of skin cancer associated with prolonged exposure to UV-A.
Dimerisation may occur between bases belonging to the same strand of DNA or between bases from different DNA strands. It produces two different types of dimers in DNA: 6-4 photoproducts (Fig. 5a) and cyclobutane pyrimidine dimers (CBP) (Fig. 5b).
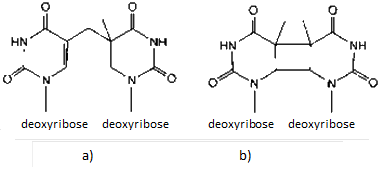
a) 6-4 photoproducts - may be 5'-T/ C- 3', 5'-C/ C- 3' or 5'-T/ Т- 3'.
b) Cyclobutane pyrimidine dimer (this is a thymine-thymine dimer).
UV light is unconditionally needed in small doses by all living creatures, as its energy is used many biochemical conversions - e.g. the synthesis of cholecalciferol (Vitamin D3) from 7-dehydrocholesterol in the skin in animals, the synthesis of serotonin in fungi and plants, etc. [
One of the most commonly used anti-UV mechanism in higher animals and in man is simple barrier protection (eumelanin barrier). The barrier properties of melanin against UV irradiation are very efficient, as over 99.9% of the absorbed dose of UV is converted to heat. UV-induced damage actually results from the residual 0.1% of the UV dose, usually UV-B. The melanin content in human skin may vary greatly, which means that different skin types exhibit different degrees of UV-sensitivity. The combined efficiency of the melanin barrier protection and the NER pathway is usually very high in healthy (repair-proficient) individuals and significant risk for carcinogenesis arises only occurs when the natural defences are faulty. The latter may be the case in repair-deficient individuals (some inherited diseases, such as xeroderma pigmentosum) or in repair-proficient individuals, if UV irradiation has been intense and/or long enough (in ageing) and/or has happened often enough so as to overcome all of the anti-UV defence mechanisms.
2.2.2. Formation of bulky adducts in DNA
Bulky adducts in DNA create steric impediments for processes involving DNA manipulation (transcription, replication). Bulky adducts getting in the way of transcription cause stalling of DNA polymerase II at the damage site and recruitment of the cell repair machinery. The presence of bulky adducts in the DNA of a dividing cell would normally cause replication arrest for assessment of the scope and degree of damage and recruitment of the repair machinery of the cell, or, the damage is deemed to be irreparable, the damaged cell would be routed to the suicide pathway.
Aromatic compounds and, specifically, polycyclic aromatic hydrocarbons (PAH) are among the commonly encountered and potent adduct-forming environmental agents. Another common adduct-forming agent is benzopyrene diol epoxide, which is produced in vivo from benzopyrene (a compound of tobacco smoke). Bulky adducts associated with benzopyrene metabolites have been implicated in the mutagenesis of the TP53 gene in smoking-related lung cancers [
Repair of regions containing bulky adducts is a complex task and may require more than one type of repair mechanism. NER is usually involved, and also repair by recombination.
2.2.3. Alkylation of exogenous origin
Among the more commonly encountered alkylating agents are the haloalkanes (dichloromethane, trichloromethane (chloroform), tetrachloromethane), alkyl sulfonates, nitrosoureas, and others. One of the most infamous warfare chemical agents, mustard gas (ypérite, LOST - widely used in World War I and II for its vesicant (blistering) properties), is an alkylating agent. Some of the most potent and widely used anticancer agents ('classic', e.g. nitrogen mustards such as cyclophosphamide or 'non-classic', such as dacarbazine) work by alkylation of DNA (see below).
2.2.4. Free radical species of exogenous origin
Free radical species, including reactive oxygen species (ROS - superoxide radicals, free hydroxyl radicals, etc.) are generated in large quantities as a result of normal cell metabolism, but they may be generated by environmental insults as well. The strength of the genotoxic action of the former is by no means inferior to that of the latter. The majority of ROS of exogenous origin result from UV irradiation, usually with UV-A (300-400nm). ROS are also generated by radiolysis of water by ionising radiation (alpha, beta or gamma).
2.2.5. DNA breaks induced by exogenous factors
Generally, it takes more intensive treatment to induce a double-strand break than a single-strand break. SSBs may result from a variety of chemical agents, long-wavelength UV and even infrared energy. Exogenously induced DSBs in DNA are usually product of high-energy electromagnetic radiation (specifically ionising radiation) and certain chemicals commonly termed as clastogens. Clastogen is an umbrella term for a variety of agents with genotoxic action which may produce double-strand breaks in DNA. Among these are, for example, commonly used chemicals such as benzene; some dyes, such as acridine yellow; some chemotherapeutic agents (radiomimetics), such as bleomycin and enediynes (e.g. neocarzinostatin); arsenic derivatives (working by increasing the amount of ROS, which, in turn, may induce double-strand breakage); some alkaloids (mimosine), and others.
Double-strand breaks may be generated directly (physical breakage of the phosphodiester bond in the DNA backbone - for example, by high-energy particles) or indirectly (e.g. by generation of free radical species, which, in turn, cause double-strand breaks). Ionising radiation may work both ways, while radiomimetic drugs usually employ the latter mechanism.
2.2.6. Thermal (infrared) damage to DNA
Under standard conditions (20°C, 100kPa) heat (infrared energy) may cause single-strand breaks or accelerate base hydrolysis. As we already saw, the loss of bases is not really significant until temperature reaches very high values, so thermal damage is only really important for prokaryotic organisms living in very hot habitats, for example, hot thermal springs, where the ambient temperature may be over 60°С. The DNA damaging effects of broadband infrared radiation is sometimes used in anticancer therapy (photothermal therapy), often together with chemotherapeutic agents [
2.2.7. DNA damage caused by microwave and radiofrequency electromagnetic radiation
Microwave radiation is electromagnetic radiation with wavelengths in the range of 25µm-1mm and frequency range between 300-3000GHz. Radiofrequency may partially overlap the microwave spectrum, but its wavelengths are generally >1mm (up to 1m) and the frequency range us under 300GHz. Microwave and radio wave energy can inflict direct DNA damage, similar to infrared damage - usually strand breaks and oxidative damage related to increased production of reactive oxygen species [
2.2.8. Tautomeric isomerism of nitrogenous bases (keto-enol and amino-imino)
Tautomerisation of nitrogenous bases is usually a product of ionising radiation. Generally, thymine and guanine in DNA are in their keto form rather than enolic from and adenine and cytosine - in amino form rather than imino form (Fig. 6).
Different tautomers may exhibit non-canonical pairing preferences. For example, thymine in its enolic from preferentially pairs with guanine and not adenine. Guanine-enol pairs with thymine. Imino-cytosine pairs preferentially with adenine and, finally, adenine in its imino form pairs with cytosine. Thus, in the next cycle of DNA replication, an A:T pair may be substituted to a G:C pair and vice versa [
Depending on whether the damage affects the sequence of the nucleotides in DNA or its structural properties, DNA damage can be further subdivided into damage associated with alterations in the information content of DNA or damage associated with structural alterations of DNA. Any of the two basic types of alterations in DNA may produce pathological phenotypes in man.
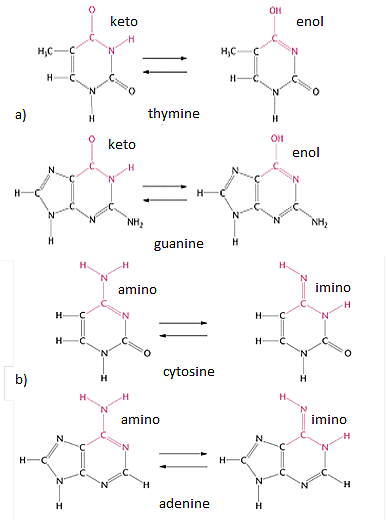
а) keto-enol; b) amino-imino.
2.3. Alterations in the information content of DNA
As DNA is a double-stranded structure and one strand is complementary to the other, the information content is always stored in two copies, the one serving as a template for synthesis of the other. If only one of these copies is intact, the information coded in the DNA sequence is fully recoverable. Problems arise only in the case where the repair of a damaged region of DNA accidentally produces a sequence which is different from the original blueprint. This may happen for a variety of reasons - from a simple base mismatch to an incorrect strand break repair. As long as the altered DNA serves only as a template for transcription, the problem is still manageable, as the functions of the damaged cell may be overtaken by other cells and differentiated cells usually have a limited lifespan. If the cell is supposed to divide, however, the altered DNA is inherited by its progeny. Changes in the information content of DNA usually do not obstruct or impede in any way the basic processes in the cell which have to do with the storage and propagation of genetic information (replication, transcription). Nevertheless, the cell carrying the genetic alteration and, in the long term, the organism, may suffer serious late consequences.
Alterations of the information content of DNA are related directly to the 'sense' of the encoded information. Substitutions of one base with another may result in synthesis of proteins with amino acid substitutions (missense mutations) or in introduction of premature stop signals, producing truncated proteins (nonsense mutations).
2.3.1. Missense mutations
The genetic code is degenerated, that is, one amino acid may be coded by more than one nucleotide triplet combinations. Therefore, a substitution of one nitrogenous base with another would not always result in a change in the information content of the coded fragment. Usually one amino acid residue is coded by at least two triplets. The molecular recognition between the third base of the mRNA codon and the first base of the tRNA anticodon is normally not very stringent due to steric effects. Triplets coding for the same amino acid often differ only in the nucleotide at position three in the triplet [
Whether the properties of the protein produced by translation of the altered DNA sequence would be drastically different from the wild type protein depends largely on the physicochemical properties of the amino acid/s involved in the substitution. At least hypothetically, substitutions between amino acid residues with similar properties would affect the properties of the resultant protein less than substitution between amino acid residues with different properties (e.g. polarity, length of the aliphatic chain, etc.). For example, a substitution of one polar amino acid residue with another polar amino acid residue would affect the properties of the resulting protein to a lesser degree than a substitution of a polar amino acid residue with a nonpolar one (and vice versa). A classic example is the most common mutation causing sickle cell anemia, an А → Т substitution in the human beta-globin gene, producing a substitution of a glutamate residue (negatively charged) with a valine residue (electrostatically neutral) in the human beta-globin protein. The mutant beta-globin chain (sickle haemoglobin, HbS) is functionally aberrant and prone to intracellular aggregation. The beta-globin aggregates cause loss of erythrocyte plasticity that may result in haemolysis and vaso-occlusion. This may occur spontaneously (without an identifiable triggering factor) or following certain triggers (high altitudes, dehydration, metabolic acidosis, etc.).
Obviously, single-nucleotide changes in DNA may affect very seriously the properties of the encoded protein. There are exceptions to this, of course. For example, in clinically healthy individuals the gene coding for the blood clotting Factor IX may exist in two alternative forms because of an A-to-G transition in exon VІ of the Factor IX gene producing a Thr-to-Ala substitution at position 148 in the protein molecule (Malmö polymorphism) [
Single-nucleotide substitutions may, albeit rarely, produce a beneficial phenotype, that is, they may augment or modify the properties of the encoded proteins so that the carrier individuals may be endowed with some selective advantage. For example, about 25% of the world population are carriers of a variant allele of the peroxisome prolife¬rator-activated receptor γ gene (PPARγ), which is associated with a decreased risk for diabetes type 2 [
Nucleotide substitutions which eventually result in substitution of one amino acid residue to another in protein-coding genes (missense mutations) may affect adversely the properties of the encoded protein with regard to their function and/or stability or may (very rarely) confer selective advantages. Most of the substitution events, however, produce no effect on the phenotype whatsoever or, at least, not any immediate effect (for example, a substitution may produce an effect which manifests only at advanced age or in response to certain triggers). Nonsense mutations and frameshift mutations may produce drastic changes in the properties and/or the expression of the resultant proteins.
2.3.2. Nonsense mutations
Nonsense mutations convert a normal triplet coding for an amino acid residue to a stop signal. This may result in premature degradation of the mRNA containing the illegitimate stop codon (therefore, no protein at all or minimal levels of synthesis) and/or in premature termination of mRNA translation, producing a truncated non-functional protein, or a protein with altered properties. In most cases this affects adversely the properties of the synthesised protein, producing a severe phenotype of genetic disease, or, in cases of somatic mutations of tumour-suppressor genes, cancerous transformation. As always, there are exceptions to the rule. The specialised literature contains descriptions of rare cases of UV-sensitivity in otherwise clinically healthy adults which were found to be homozygous carriers of nonsense mutations in ERCC6, a gene with crucial role in DNA repair [
Inherited nonsense mutations usually produce severe phenotypes of genetic disease. It is only to be expected, as they are usually associated with complete lack or a severe deficiency of an important protein. Nonsense mutations are among the most common molecular defects associated with cystic fibrosis (CF), Duchenne/Becker muscular dystrophy (DMD/BMD), beta-thalassemia, Tay-Sachs disease, etc. There is, however, a rare natural workaround mechanism for compensating the deficiency (at least partially). It is not unusual to detect small amounts of normal-sized protein in patients with diseases and conditions due to nonsense mutations, albeit the levels of the deficient protein are usually low or very low. This is believed to result from read-through translation of the defective mRNA sequence which ignores the premature termination codon. Read-through translation may produce an unusually mild phenotype [
Ataluren has shown effectiveness in clinical trials phase 2a for treatment of DMD/BMD as well [
A Phase 2 trial of Ataluren for patients with methylmalonic aciduria was initiated in 2010, but no results have been published so far.
Ataluren and the aminoglycoside Geneticin have been assessed as termination codon-ignoring agents in other genetic diseases associated with nonsense mutations, such as limb-girdle muscular dystrophy and some types of peroxisome biogenesis disorders [
It is notable that the read-through effect of Ataluren is not equally efficient on all termination codons. The suppression of the reading of termination codons is most pronounced for the UGA termination codon (opal) [
2.3.3. Frameshift mutations
Frameshift mutations comprise all insertions and deletions of short nucleotide sequences (up to 30-40 bp), the length of which is not a multiple of 3. The open reading frame (ORF) is disrupted beyond the mutation site; therefore, the information content of the nucleic acid would change from this point onward. The properties and/or the stability of the resulting transcript and the encoded protein may change drastically, even if the deletion or insertion is only one or two bp in length.
The deleterious effect of frameshift mutations is related, on the one hand, to the risk of altering the amino acid sequence beyond the site of the mutation, as triplet code may read very differently from the wild type sequence after the insertion or the deletion have occurred (Fig. 7).
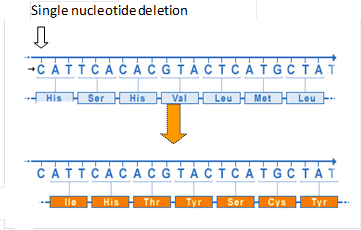
Reading frames other than the ORF may contain one or more stop codons; therefore, once the frameshift has occurred, the risk for encountering a premature stop codon increases along the length of the mutated sequence. The translation of the mRNA beyond the mutation site is usually terminated early, producing a truncated and/or instable protein.
Frameshift mutations in many human genes are associated with risk of inherited disease. Usually, the defect is more severe in cases of small deletions and insertions of fragments whose length is not a multiple of 3 than of larger (sometimes, much larger) fragments with length that is a multiple of 3. For example, most deletions in the human F8 gene (even very small ones) usually produce a severe bleeding phenotype with virtually undetectable levels of circulating Factor VIII protein. An exception is the deletion of exon 22 (156 bp, that is, multiple of 3) [
There is one particular frameshift mutation in a human gene which brings more than one selective advantages to its carriers, (especially homozygous carriers). It is commonly seen throughout Europe (though areas of high and low prevalence exist) and rare in Africa and Asia. The mutation in question is a 32 bp (that is, frameshift) deletion in the human chemokine receptor 5 gene. The homozygous carriership of the CCR5del32 has been known for almost 20 years to be associated with delayed progression from HIV carriership to overt HIV-related disease in HIV-infected individuals [
A novel therapeutic approach has been developed for human diseases due to mutations disrupting the reading frame, too. This is exon-skipping therapy (EST), which works basically by eliminating the exon containing the molecular defect from the polypeptide chain (adjacent exon/s may be removed as well). Thus, the resultant protein may be truncated but at least some of its functions may be restored, provided that the remaining exons are spliced in-frame. The EST approach is applicable to nonsense mutations as well. As in read-through therapy of (see above), the main goal of exon skipping for treatment of disease is not cure, but, rather, an amelioration of the disease phenotype. If we use DMD/BMD as example again, frameshift mutations usually produce a severe DMD phenotype with very low or undetectable levels of dystrophin while deletions of one or more exons usually produce the milder Becker phenotype (as long as the remaining exons are spliced in-frame), where dystrophin is present but its functionality is altered. Therefore, skipping one or two exons may produce a phenotype modification, shifting the clinical manifestations from the more severe form (DMD) towards the milder phenotype (BMD). The latter may make all the difference to the patients and their families, as DMD patients usually have earlier onset of the disease, more rapid progression of muscle wasting and shorter life expectancy (usually below 25 years), while BMD patients usually exhibit a later onset, slower disease progression and may have life expectancy well into their 40-ties and beyond. The therapeutic agents used in EST are antisense RNA oligomers which work by switching splice sites in mRNA [
2.4. Structural alterations in DNA
Examples for structural DNA alterations are single-strand or double-strand breaks, genomic rearrangements, abasic sites, covalent linkages between nitrogenous bases in the same strand or different strands, DNA-protein crosslinks, Z-form of DNA, triple-stranded DNA, etc. Base mismatches (before they are converted to alterations in the 'sense' of the sequence) are also believed to constitute structural alterations in DNA.
Structural alterations in DNA create steric impediments to DNA replication and/or transcription. Copying of altered DNA may be temporarily stalled or arrested altogether. DNA templates carrying structural alterations may be copied incorrectly, efficiently producing a heritable mutation. Also, alterations in the information content of DNA may result in structural changes. For example, expansion of repeats in DNA, which is, essentially, an alteration of its information content, may increase the probability of forming alternative structures in DNA, such as triple-helix regions [
2.4.1. Genomic rearrangements
Rearrangements are not uncommon events in eukaryotic genomes. They may occur in the germline, producing abnormal karyotypes (with or without associated disease) or in somatic cells (which may result in cancer). For example, a common cytogenetic abnormality found in the majority of patients with chronic myelogenous leukemia (CML) is the Philadelphia chromosome [
Many of the hereditary cancer-prone syndromes (Werner syndrome, Bloom syndrome, and others) are associated with increased rate of genome rearrangements. Repair by non-homologous end joining may occasionally result in genomic rearrangements.
2.4.2. Gene conversion
The template copying mechanism of DNA repair has its drawbacks, as the simple recovery of the lost or altered DNA sequence is not sufficient. The disadvantages, however, surface only rarely, when the template strand has also been altered so that it became different from the 'correct' blueprint. Gene conversion occurs as a result of homologous recombination between closely related DNA sequences. Homologous recombination (HR) is a normal cellular mechanism for repair of double-strand breaks. Gene conversion employs one DNA sequence as substrate for repair by recombination with another DNA sequence, effectively substituting the latter with the former. If the DNA sequence used as template carries alterations which make it different from the sequence which is under repair, the alterations in the template may be permanently introduced in the repaired sequence. Gene conversion has the potential to introduce alterations in the information content of DNA (exchanging one genomic sequence for another) as well as structural alterations (as it causes movement of fragments of DNA from one site to another).
Genetic disease may result from gene conversion. For example, many cases of congenital adrenal hyperplasia due to 21-hydroxylase deficiency are associated with gene conversion between the functional CYP21A2 gene copy and a closely related, highly homologous pseudogene located on the same chromosome [
2.4.3. Z-form of DNA
The right-handed double-strand B-form of DNA is the most common DNA form found in living cells [
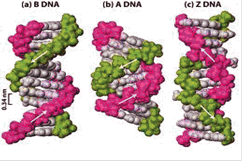
Apart from the helix sense (the direction of the winding of the spiral), Z-DNA exhibits other characteristics different from the properties of the A- and B-forms. For example, Z-DNA is 'thinner' with regard to its helix diameter (with A-DNA being the broadest and B-DNA of intermediate diameter); the number of base pairs per helix turn are 12 in the Z-form compared to the 10-11 turns of the other two forms; the base pairs are tilted at an angle of 9° to the helix axis (compared with the 19° of the A form and the 1° of the B-form). The topology of the grooves is also different between different forms of DNA, as Z-DNA has a flat major groove and a narrow deep minor groove while the other two forms generally have a shallow minor groove and a deep major groove [
Z-DNA may arise in vivo as a product of the B-form by relaxation of the torsion strain (supercoiling) of B-DNA. Supercoiling often occurs during in vivo DNA manipulation. For example, the progress of the RNA polymerases along the length of the DNA molecule during transcription results in negative supercoiling [
Some of the enzymatic activities which act in repair of modified bases (О6-methylguanine methyltransferase) may not recognise modified nucleotides, buried in a DNA region in Z-form [
2.4.4. Triple-helix DNA
Triple-helix (triple-strand) DNA arises when a single DNA strand binds by hydrogen bonds (but not by Watson-Crick base pairing) to a preformed B-form DNA duplex [
The relative role of alterations of information content of DNA and structural alterations in DNA is difficult to assess as a factor in human disease. Some damaging agents cause DNA damage predominantly of the one or the other type [
2.5. Application of DNA damaging agents in anticancer therapy
Paracelsus (1493-1541)
Modern anticancer therapy is largely based on the use of substances and/or agents acting by holding back or slowing down the proliferation of cancer cells. In most cases antitumour therapeutics are typical DNA-damaging agents, that is, they work by creating replication blocks, inhibiting the 'untangling' of DNA during nuclear transactions, introducing adducts and breaks in DNA, etc.
The capacity for infliction of DNA damage (genotoxicity) of various chemical and physical agents has been used in the treatment of human cancer even before the associated mechanisms of damage detection and repair and damage-induced cell death were made clear. The first ever chemotherapeutic drug used to treat malignant disease (lymphoma) in 1942 was an alkylating agent, a mustard gas derivative - namely, nitrogen mustard [
The consequences of the DNA damage inflicted by DNA damaging agents are suffered by all cells of the organism. Since the cells who divide at the fastest rate tend to be the most affected by genotoxic treatments, the cell types that receive the major impact of genotoxic damage are the cancer cells (which are, after all, the intended target), but also healthy cells who normally have rapid turnover, such as epidermal cells (skin, mucosa and skin appendages such as hair) and haematopoietic cells. Usually, after the acute genotoxic influence is over, the functions of the affected cell populations are restored over time. Also, the toxic effects of antitumour therapy are usually dose limited, unless other factors (e.g. carriership of certain DNA polymorphisms, long-term effects of some chemical agents, such as compounds in tobacco smoke, etc.) are present which may precipitate increased toxicity or resistance to therapy.
Anticancer agents with genotoxic properties may be of chemical (e.g. cytostatic medication) or physical nature (e.g. ionising radiation). Below we present the properties of the major groups of modern anticancer drugs that work by inflicting DNA damage in tumour cells.
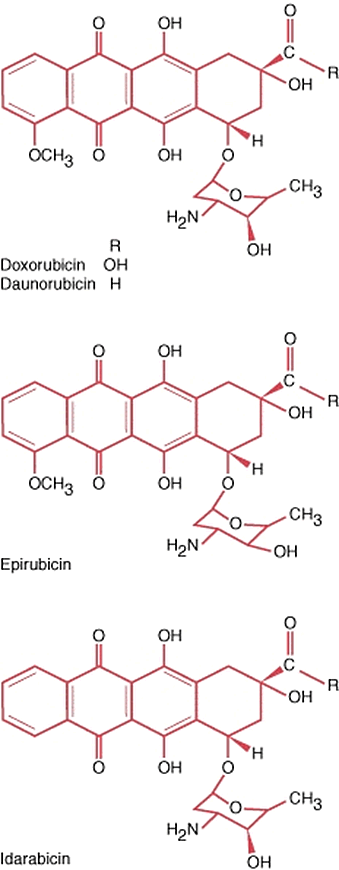
2.5.1. Anthracycline and anthracycline derivatives
Anthracyclines (Fig. 9) are fermentation products of Actinobacteria of the genus Streptomyces. Historically, the first proof of the antitumour capacity of anthracyclines appeared in the late 60-ties of the XX century for daunomycin, isolated from Streptomyces peucetius var. caesius [
The antitumour properties of anthracyclines are exerted via more than one mechanism [
Anthracyclines are often used as first-line drugs in the treatment of breast cancer and other carcinomas, leukemias, soft tissue sarcomas, etc. They may be used alone or in combination with other antitumour agents, such as vincristine and bleomycin. Doxorubicin has the broadest spectrum of activity in the anthracycline group so far.
The overall response rate for anthracyclines (defined as the proportion of patients in which the tumour shrinks in size or becomes undetectable by routine methods) is between 40 and 50%, depending on the type, size and site of the tumour. The response rate for anthracyclines is comparable only to the rate achieved with platinum derivatives (see below). All other contemporarily used chemotherapeutics are less effective in terms of their overall response rate, unless they are used in combined therapeutic regimens.
The most frequently seen adverse effect of anthracycline treatment is inhibition of haematopoiesis, manifesting as leuko- and neutropenia that may become clinically significant. Another serious adverse effect is cardiotoxicity, which may result in acute cardiac effects or congestive heart failure. Anthracyclines also produce transitory but universal epilation, mucositis and gastrointestinal disturbances (nausea, vomiting and diarrhoea).
2.5.2. Anthracenediones
These are anthracycline analogues characterised by decreased (but still significant) cardiac toxicity. Mitoxantrone (anthracenedione) (Fig. 10) is an inhibitor of DNA topoisomerase II, like the generic anthracycline. It works by introducing double-strand breaks in DNA which serve as a potent signal for induction of cell cycle arrest. Mitoxantrone is often used in combination therapy with cytosine arabinoside (cytarabine), a pyrimidine analogue (see below).
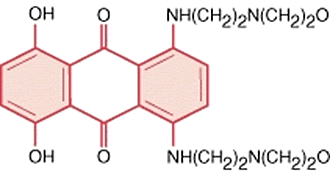
Mitoxanthrone is less effective in forming free radicals that the anthracyclines and is has lower DNA intercalating properties [
2.5.3. Alkylating agents
This group of anticancer chemotherapeutics is rather diverse, including the so-called 'classic' alkylating drugs, such as cyclophosphamide, ifosfamide, busulfan, melphalan, chlorambucil (Fig. 11), etc., as well as some 'non-classic' such as altretamine, procarbazine and dacarbazine (Fig. 12). The classic alkylating drugs work by direct alkylation of DNA bases (usually on a guanosine residue). The newer 'non-classic' alkylators are metabolised by a variety of pathways, eventually producing a reactive methyl group methylating DNA [
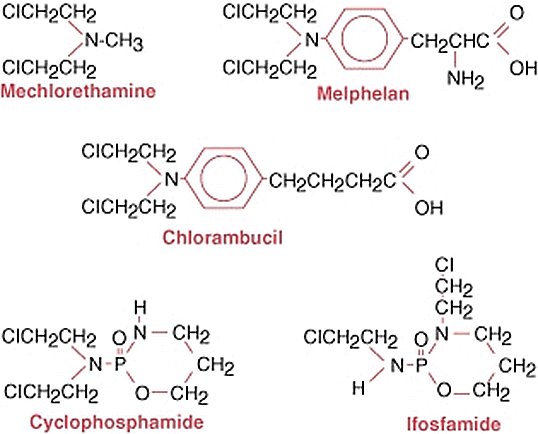
Nitrogen mustards constitute a major group in classic alkylating agents. Among these, oxazaphosphorines (cyclophosphamide, ifosfamide, trofosfamide) are most commonly used, closely followed by another nitrogen mustard, melphalan. Cyclophosphamide is often used as a first-line treatment of leukemia and lymphoma, and also in chemotherapy regimens for some solid tumours such as breast cancer. The nitrogen mustards are metabolised in the liver, producing phosphoramide mustard, which is the active metabolite. The mechanism of action is based on the capacity of the phosphoramide mustard to induce crosslinks in DNA, between deoxyguanosine residues in the same strand (interstrand) as well as between strands (intrastrand crosslinks) [
Another commonly uses therapeutic application of cyclophosphamide is immunosuppressive therapy. Like other cytostatic drugs used as immunosuppressive agents (e.g. the mentioned above mitoxanthrone), cyclophosphamide increases the long-term risk for cancer [
The alkylsulfonate busulfan used to be a first-line therapy for certain types of leukemia, but currently is more and more often substituted with the tyrosine kinase drug Imatinib. Busulfan treatment is, however, less expensive; therefore, it may still be prescribed whenever cost of treatment is critically important.
Non-classic alkylating agents are usually used as a part of a combination regimen and are not first-line treatment choices, unless in a combination with other anticancer drugs. Single-agent use of non-classic alkylators has been shown to be associated with low complete response rates and short-lived beneficial effects [
2.5.4. Agents producing bulky adducts in DNA
Typical representatives of this group are platinum-based anticancer drugs - cisplatin (the first to be identified and developed), carboplatin, oxaliplatin, picoplatin, triplatin, and others. As with many significant discoveries, the identification of the anticancer properties of platinum compounds was purely accidental. In 1965, Barnett Rosenberg, a chemist working at the time in the Biophysics department of the Michigan State University, was studying the effects of electric fields on bacterial growth. He and his co-workers repeatedly observed that bacterial cells placed in electric current would stop dividing, but grew in size, and made the conclusion that there was an unknown compound, possibly an electrolysis product that inhibited their division [
Platinum compounds are coordination complexes of platinum ion with four or six ligands, with a square planar or hexahedral configuration, respectively (Fig. 13).
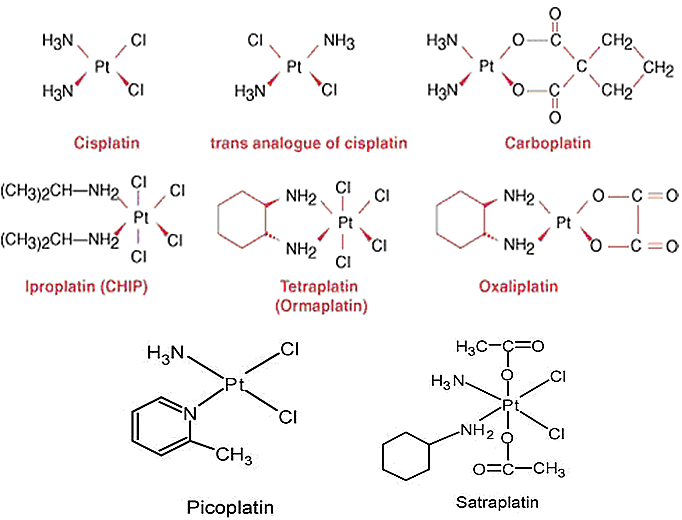
Classic platinum compounds (first generation - cisplatin, second generation - carboplatin, third generation - oxaliplatin) and some of the newer platinum-based drugs (e.g. picoplatin, considered to be a third generation drug) are platinum (II), while others (e.g. satraplatin, the only orally administered platinum derivative) are platinum (IV) complexes. Due to their unique structure, platinum anticancer agents are not metabolised or biodegraded, as most anticancer drugs are. To yield a reactive moiety capable of interaction with DNA within the cell, the chlorine atoms attached to the platinum ion in the centre of the complex must be removed. This is not implemented by catalytic events, however, but by spontaneous displacement reactions by electron donors, e.g. water molecules (aqua ligands), sulphur-containing groups (e.g. from proteins - e.g. glutathionе, metallothionein, etc. and amino groups - cysteine, methionine). The aqua ligands may then be exchanged with nitrogen atoms from nitrogenous bases in DNA (commonly, N7 from closely located guanines or, more rarely, N7 or N1 of adenines), forming bulky adducts in DNA [
Cisplatin is fairly unstable in vivo, the half-life being between 20 and 30 minutes [
Platinum compounds are among the most commonly used and most efficient anticancer therapeutics, with a response rate of 25-35% when used as a single agent and over 50% (in some cases, up to 70%) when used in combination with other agents [
Another group of chemotherapeutic drugs acting by inducing crosslinks in DNA are the aziridine derivatives, among which prominent are mitomycin C and thioTEPA. Mitomycin C has specificity for guanine residues at CpG dinucleotides in DNA [
2.5.5. Antimetabolites (nucleotide analogues)
These are modified purine analogues of 'regular' nucleotides in DNA or compounds that inhibit the synthesis of 'regular' nucleotides. Example of the latter is the most commonly used antimetabolite drug, methotrexate. It works by inhibition of dihydrofolate reductase (DHFR), an enzyme catalysing the conversion of dihydrofolate to tetrahydrofolate, which is needed for the biosynthesis of thymidine and the purine nucleotides [
Some antimetabolite drugs (including methotrexate), are chemically similar to folic acid (antifolates). They usually bind to and inhibit the activity of dihydrofolate reductase. Pemetrexed is an antifolate agent capable of inhibiting more than one enzymatic activity needed for the synthesis of nucleic acids - specifically, DHFR, thymidilate synthase, and the aminoimidazole carboxamide ribonucleotide formyltransferase (AICART) [
The most popularly used pyrimidine analogues are cytarabine, gemcitabine, 5-fluorouracil, capecitabine (metabolised to 5-fluorouracil), and others. Gemcitabine also works as inhibitor of ribonucleotide reductase (RNR, the enzymatic activity catalysing the conversion of ribonucleotides to deoxyribonucleotides [reviewed in
Some antimetabolite drugs (azathioprine, methotrexate) are routinely used in immunosuppressive regimens. Other antimetabolite drugs (trimethoprim, pyrimethamine, proguanil and others) may be used in treatment of infectious disease caused by protozoa (malaria, toxoplasmosis, etc.) [reviewed in
Most nucleotide analogues exhibit long-term carcinogenic effects [
2.5.6. Chemotherapeutics introducing double-strand breaks in DNA (radiomimetics)
Radiomimetics are a group of drugs whose genotoxic effects resemble the DNA damage caused by ionising radiation (double-strand breaks). This group of anticancer drugs generally works by stimulation of the intracellular production of ROS, which, in turn, induce double-strand breaks in DNA [
Sensitivity to radiomimetics and, for that matter, to ionising radiation, is observed in several inherited disorders, such as ataxia-telangiectasia and Nijmegen breakage syndrome [
Acknowledgеments
This research was supported by Grant No. DFNI-B01/2 at the National Science Fund, Ministry of Education and Science of Republic of Bulgaria.
References
- Lewin B. Genes VIII. Prentice-Hall, Inc., Upper Saddle River, NJ 07458, USA, 2003.
- Harper JV, Anderson JA, O'Neill P. Radiation induced DNA DSBs: Contribution from stalled replication forks? DNA Repair (Amst). 2010 Aug 5;9(8):907-13.
- Deshmukh PS, Megha K, Banerjee BD, Ahmed RS, Chandna S, Abegaonkar MP, Tripathi AK. Detection of Low Level Microwave Radiation Induced Deoxyribonucleic Acid Damage Vis-à-vis Genotoxicity in Brain of Fischer Rats. Toxicol Int. 2013 Jan;20(1):19-24.
- Holme JA, Søderlund EJ. Modulation of genotoxic and cytotoxic effects of aromatic amines in monolayers of rat hepatocytes. Cell Biol Toxicol. 1984 Oct;1(1):95-110.
- Roy Chowdhury A, Bakshi R, Wang J, Yildirir G, Liu B, Pappas-Brown V, Tolun G, Griffith JD, Shapiro TA, Jensen RE, Englund PT. The killing of African trypanosomes by ethidium bromide. PLoS Pathog. 2010 Dec 16;6(12):e1001226.
- Juzeniene A, Stokke KT, Thune P, Moan J. Pilot study of folate status in healthy volunteers and in patients with psoriasis before and after UV exposure. J Photochem Photobiol B. 2010 Nov 3;101(2):111-6.
- El-Saie LT, Rabie AR, Kamel MI, Seddeik AK, Elsaie ML. Effect of narrowband ultraviolet B phototherapy on serum folic acid levels in patients with psoriasis. Lasers Med Sci. 2011 Jul;26(4):481-5.
- Blatter BM, Hermens R, Bakker M, Roeleveld N, Verbeek AL, Zielhuis GA. Paternal occupational exposure around conception and spina bifida in offspring. Am J Ind Med. 1997 Sep;32(3):283-91.
- Ames BN, Lee FD, Durston WE. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc Natl Acad Sci U S A. 1973 Mar;70(3):782-6.
- McCann J, Spingarn NE, Kobori J, Ames BN. Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids. Proc Natl Acad Sci U S A. 1975 Mar;72(3):979-83.
- Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991 Jun 15;51(12):3075-9.
- Chen X, Zuo S, Kelman Z, O'Donnell M, Hurwitz J, Goodman MF. Fidelity of eucaryotic DNA polymerase delta holoenzyme from Schizosaccharomyces pombe. J Biol Chem. 2000 Jun 9;275(23):17677-82.
- Kunkel TA. DNA replication fidelity. J Biol Chem. 2004 Apr 23;279(17):16895-8.
- McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008 Jan;18(1):148-61.
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010 Mar 12;37(5):714-27.
- Huang JC, Sancar A. Determination of minimum substrate size for human excinuclease. J Biol Chem. 1994 Jul 22;269(29):19034-40.
- Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000 Apr 27;404(6781):1014-8.
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000 Aug 31;406(6799):1015-9.
- Rattray AJ, Strathern JN. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu Rev Genet. 2003;37:31-66.
- Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007 Feb 23;25(4):601-14.
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009 Mar;73(1):134-54.
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998 Apr;148(4):1667-86.
- Nachman MW1, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000 Sep;156(1):297-304.
- Bensasson D, Feldman MW, Petrov DA. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J Mol Evol. 2003 Sep;57(3):343-54.
- Vitkup D, Sander C, Church GM. The amino-acid mutational spectrum of human genetic disease. Genome Biol. 2003;4(11):R72.
- Wu H, Ma BG, Zhao JT, Zhang HY. How similar are amino acid mutations in human genetic diseases and evolution. Biochem Biophys Res Commun. 2007 Oct 19;362(2):233-7.
- Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683-706.
- Moseley BE, Mattingly A. Repair of irradiation transforming deoxyribonucleic acid in wild type and a radiation-sensitive mutant of Micrococcus radiodurans. J Bacteriol. 1971 Mar;105(3):976-83.
- Saffary R, Nandakumar R, Spencer D, Robb FT, Davila JM, Swartz M, Ofman L, Thomas RJ, DiRuggiero J. Microbial survival of space vacuum and extreme ultraviolet irradiation: strain isolation and analysis during a rocket flight. FEMS Microbiol Lett. 2002 Sep 24;215(1):163-8.
- Abrevaya XC, Paulino-Lima IG, Galante D, Rodrigues F, Mauas PJ, Cortón E, Lage Cde A. Comparative survival analysis of Deinococcus radiodurans and the haloarchaea Natrialba magadii and Haloferax volcanii exposed to vacuum ultraviolet irradiation. Astrobiology. 2011 Dec;11(10):1034-40.
- Horikawa DD, Sakashita T, Katagiri C, Watanabe M, Kikawada T, Nakahara Y, Hamada N, Wada S, Funayama T, Higashi S, Kobayashi Y, Okuda T, Kuwabara M. Radiation tolerance in the tardigrade Milnesium tardigradum. Int J Radiat Biol. 2006 Dec;82(12):843-8.
- Ivanov VK, Chekin SIu, Kashcheev VV, Maksiutov MA, Tumanov KA, Tsyb AF. Mortality among the liquidators of the Chernobyl accident: dose dependences and groups of the potential risk. Radiats Biol Radioecol. 2011 Jan-Feb;51(1):41-8.
- McMurray BJ. 1976 Hanford americium exposure incident: accident description. Health Phys. 1983 Oct;45(4):847-53.
- Toohey RE, Kathren RL. Overview and dosimetry of the Hanford Americium accident case. Health Phys. 1995 Sep;69(3):310-7.
- Thiviyanathan V, Somasunderam A, Volk DE, Hazra TK, Mitra S, Gorenstein DG. Base-pairing properties of the oxidized cytosine derivative, 5-hydroxy uracil. Biochem Biophys Res Commun. 2008 Feb 15;366(3):752-7.
- Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res. 1993 Jan;285(1):61-7.
- Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005 Nov 27;33(20):e176.
- Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1642-6.
- Costello JF, Plass C. Methylation matters. J Med Genet. 2001 May;38(5):285-303.
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001 Apr 15;61(8):3225-9.
- Sekigawa I, Kawasaki M, Ogasawara H, Kaneda K, Kaneko H, Takasaki Y, Ogawa H. DNA methylation: its contribution to systemic lupus erythematosus. Clin Exp Med. 2006 Oct;6(3):99-106.
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993 Apr 22;362(6422):709-15.
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610-8.
- Lindahl T, Karlström O. Heat-induced depyrimidination of deoxyribonucleic acid in neutral solution. Biochemistry. 1973 Dec 4;12(25):5151-4.
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis, 2nd ed., ASM Press, ISBN 1-55581-319-4.
- Lindahl T, Ljungquist S. Apurinic and apyrimidinic sites in DNA. Basic Life Sci. 1975;5A:31-8.
- Loechler EL, Green CL, Essigmann J. In vivo mutagenesis by 06-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271-5.
- Preston BD, Singer B, Loeb LA. Mutagenic potential of 04-methylthymine in vivo determined by an enzymatic approach to site-specific mutagenesis. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8501-5.
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009 Dec;1(2):239-59.
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999 Oct;23(2):185-8..
- Watson P, Black G, Ramsden S, Barrow M, Super M, Kerr B, Clayton-Smith J. Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. J Med Genet. 2001 Apr;38(4):224-8.
- van Loon B, Markkanen E, Hübscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst). 2010 Jun 4;9(6):604-16.
- Barbin A, Wang R, O'Connor PJ, Elder RH. Increased formation and persistence of 1,N(6)-ethenoadenine in DNA is not associated with higher susceptibility to carcinogenesis in alkylpurine-DNA-N-glycosylase knockout mice treated with vinyl carbamate. Cancer Res. 2003 Nov 15;63(22):7699-703.
- Gale KC, Osheroff N. Intrinsic intermolecular DNA ligation activity of eukaryotic topoisomerase II. Potential roles in recombination. J Biol Chem. 1992 Jun 15;267(17):12090-7.
- Shuman S. Polynucleotide ligase activity of eukaryotic topoisomerase I. Mol Cell. 1998 Apr;1(5):741-8.
- Nicolas N, Moshous D, Cavazzana-Calvo M, Papadopoulo D, de Chasseval R, Le Deist F, Fischer A, de Villartay JP. A human severe combined immunodeficiency (SCID) condition with increased sensitivity to ionizing radiations and impaired V(D)J rearrangements defines a new DNA recombination/repair deficiency. J Exp Med. 1998 Aug 17;188(4):627-34.
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001 Apr 20;105(2):177-86.
- Barlow C, Dennery PA, Shigenaga MK, Smith MA, Morrow JD, Roberts LJ 2nd, Wynshaw-Boris A, Levine RL. Loss of the ataxia-telangiectasia gene product causes oxidative damage in target organs. Proc Natl Acad Sci U S A. 1999 Aug 17;96(17):9915-9.
- Stern N, Hochman A, Zemach N, Weizman N, Hammel I, Shiloh Y, Rotman G, Barzilai A. Accumulation of DNA damage and reduced levels of nicotine adenine dinucleotide in the brains of Atm-deficient mice. J Biol Chem. 2002 Jan 4;277(1):602-8.
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanová E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998 May 1;93(3):467-76.
- Fraikin GY, Strakhovskaya MG, Ivanova EV, Rubin AB. Near-UV activation of enzymatic conversion of 5-hydroxytryptophan to serotonin. Photochem Photobiol. 1989 Apr;49(4):475-7.
- Gambichler T, Bader A, Vojvodic M, Avermaete A, Schenk M, Altmeyer P, Hoffmann K. Plasma levels of opioid peptides after sunbed exposures. Br J Dermatol. 2002 Dec;147(6):1207-11.
- Iyengar B. The melanocyte photosensory system in the human skin. Springerplus. 2013 Apr 12;2(1):158.
- Atillasoy ES, Seykora JT, Soballe PW, Elenitsas R, Nesbit M, Elder DE, Montone KT, Sauter E, Herlyn M. UVB induces atypical melanocytic lesions and melanoma in human skin. Am J Pathol. 1998 May;152(5):1179-86.
- Bennett DC. Ultraviolet wavebands and melanoma initiation. Pigment Cell Melanoma Res. 2008 Oct;21(5):520-4.
- Denissenko MF, Pao A, Pfeifer GP, Tang M. Slow repair of bulky DNA adducts along the nontranscribed strand of the human p53 gene may explain the strand bias of transversion mutations in cancers. Oncogene. 1998 Mar 12;16(10):1241-7.
- Tretyakova N, Matter B, Jones R, Shallop A. Formation of benzo[a]pyrene diol epoxide-DNA adducts at specific guanines within K-ras and p53 gene sequences: stable isotope-labeling mass spectrometry approach. Biochemistry. 2002 Jul 30;41(30):9535-44.
- Zaludová R, Zákovská A, Kasparková J, Balcarová Z, Kleinwächter V, Vrána O, Farrell N, Brabec V. DNA interactions of bifunctional dinuclear platinum(II) antitumor agents. Eur J Biochem. 1997 Jun 1;246(2):508-17.
- Stiborová M, Sejbal J, Borek-Dohalská L, Aimová D, Poljaková J, Forsterová K, Rupertová M, Wiesner J, Hudecek J, Wiessler M, Frei E. The anticancer drug ellipticine forms covalent DNA adducts, mediated by human cytochromes P450, through metabolism to 13-hydroxyellipticine and ellipticine N2-oxide. Cancer Res. 2004 Nov 15;64(22):8374-80.
- Laib RJ, Stöckle G, Bolt HM, Kunz W. Vinyl chloride and trichloroethylene: comparison of alkylating effects of metabolites and induction of preneoplastic enzyme deficiencies in rat liver. J Cancer Res Clin Oncol. 1979 Jun 8;94(2):139-47.
- Bartsch H, Malaveille C, Barbin A, Planche G. Mutagenic and alkylating metabolites of halo-ethylenes, chlorobutadienes and dichlorobutenes produced by rodent or human liver tissues. Evidence for oxirane formation by P450-linked microsomal mono-oxygenases. Arch Toxicol. 1979 Feb 23;41(4):249-77.
- Bolt HM. Vinyl chloride-a classical industrial toxicant of new interest. Crit Rev Toxicol. 2005 Apr-May;35(4):307-23.
- You J, Zhang R, Xiong C, Zhong M, Melancon M, Gupta S, Nick AM, Sood AK, Li C. Effective photothermal chemotherapy using doxorubicin-loaded gold nanospheres that target EphB4 receptors in tumors. Cancer Res. 2012 Sep 15;72(18):4777-86.
- Chang HY, Shih MH, Huang HC, Tsai SR, Juan HF, Lee SC. Middle infrared radiation induces G2/M cell cycle arrest in A549 lung cancer cells. PLoS One. 2013;8(1):e54117.
- Liu C, Duan W, Xu S, Chen C, He M, Zhang L, Yu Z, Zhou Z. Exposure to 1800 MHz radiofrequency electromagnetic radiation induces oxidative DNA base damage in a mouse spermatocyte-derived cell line. Toxicol Lett. 2013 Mar 27;218(1):2-9.
- Shahin S, Singh VP, Shukla RK, Dhawan A, Gangwar RK, Singh SP, Chaturvedi CM. 2.45 GHz microwave irradiation-induced oxidative stress affects implantation or pregnancy in mice, Mus musculus. Appl Biochem Biotechnol. 2013 Mar;169(5):1727-51.
- Klug WS, Cummings MR. Concepts of Genetics, 5th edition. Upper Saddle River, N.J.:Prentice-Hall, 1997.
- Crick FH. Codon-anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548-55.
- Graham JB, Lubahn DB, Lord ST, Kirshtein J, Nilsson IM, Wallmark A, Ljung R, Frazier LD, Ware JL, Lin SW, Stafford DW, Bosco J. The Malmö polymorphism of coagulation factor IX, an immunologic polymorphism due to dimorphism of residue 148 that is in linkage disequilibrium with two other F.IX polymorphisms. Am J Hum Genet. 1988 Apr;42(4):573-80.
- Stumvoll M, Häring H. The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism. Diabetes. 2002 Aug;51(8):2341-7.
- Fujiwara Y, Ichihashi M, Kano Y, Goto K, Shimizu K. A new human photosensitive subject with a defect in the recovery of DNA synthesis after ultraviolet-light irradiation. J Invest Dermatol. 1981 Sep;77(3):256-63.
- Miyauchi-Hashimoto H, Akaeda T, Maihara T, Ikenaga M, Horio T. Cockayne syndrome without typical clinical manifestations including neurologic abnormalities. J Am Acad Dermatol. 1998 Oct;39(4 Pt 1):565-70.
- Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M, Tanaka K. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci U S A. 2004 Oct 26;101(43):15410-5.
- Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997 Jan 17;272(3):1885-90.
- Guzder SN, Habraken Y, Sung P, Prakash L, Prakash S. RAD26, the yeast homolog of human Cockayne's syndrome group B gene, encodes a DNA-dependent ATPase. J Biol Chem. 1996 Aug 2;271(31):18314-7.
- Saviozzi S, Saluto A, Taylor AM, Last JI, Trebini F, Paradiso MC, Grosso E, Funaro A, Ponzio G, Migone N, Brusco A. A late onset variant of ataxia-telangiectasia with a compound heterozygous genotype, A8030G/7481insA. J Med Genet. 2002 Jan;39(1):57-61.
- Vissing J, Duno M, Schwartz M, Haller RG. Splice mutations preserve myophosphorylase activity that ameliorates the phenotype in McArdle disease. Brain. 2009 Jun;132(Pt 6):1545-52.
- Aurino S, Nigro V. Readthrough strategies for stop codons in Duchenne muscular dystrophy. Acta Myol. 2006 Jun;25(1):5-12.
- Malik V, Rodino-Klapac LR, Viollet L, Mendell JR. Aminoglycoside-induced mutation suppression (stop codon readthrough) as a therapeutic strategy for Duchenne muscular dystrophy. Ther Adv Neurol Disord. 2010 Nov;3(6):379-89.
- Sermet-Gaudelus I, Boeck KD, Casimir GJ, Vermeulen F, Leal T, Mogenet A, Roussel D, Fritsch J, Hanssens L, Hirawat S, Miller NL, Constantine S, Reha A, Ajayi T, Elfring GL, Miller LL. Ataluren (PTC124) induces cystic fibrosis transmembrane conductance regulator protein expression and activity in children with nonsense mutation cystic fibrosis. Am J Respir Crit Care Med. 2010 Nov 15;182(10):1262-72.
- Wilschanski M, Miller LL, Shoseyov D, Blau H, Rivlin J, Aviram M, Cohen M, Armoni S, Yaakov Y, Pugatsch T, Cohen-Cymberknoh M, Miller NL, Reha A, Northcutt VJ, Hirawat S, Donnelly K, Elfring GL, Ajayi T, Kerem E. Chronic ataluren (PTC124) treatment of nonsense mutation cystic fibrosis. Eur Respir J. 2011 Jul;38(1):59-69.
- Finkel RS, Flanigan KM, Wong B, Bönnemann C, Sampson J, Sweeney HL, Reha A, Northcutt VJ, Elfring G, Barth J, Peltz SW. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS One. 2013 Dec 11;8(12):e81302.
- Wang B, Yang Z, Brisson BK, Feng H, Zhang Z, Welch EM, Peltz SW, Barton ER, Brown RH Jr, Sweeney HL. Membrane blebbing as an assessment of functional rescue of dysferlin-deficient human myotubes via nonsense suppression. J Appl Physiol (1985). 2010 Sep;109(3):901-5.
- Dranchak PK, Di Pietro E, Snowden A, Oesch N, Braverman NE, Steinberg SJ, Hacia JG. Nonsense suppressor therapies rescue peroxisome lipid metabolism and assembly in cells from patients with specific PEX gene mutations. J Cell Biochem. 2011 May;112(5):1250-8.
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, Wilde RG, Karp G, Takasugi J, Chen G, Jones S, Ren H, Moon YC, Corson D, Turpoff AA, Campbell JA, Conn MM, Khan A, Almstead NG, Hedrick J, Mollin A, Risher N, Weetall M, Yeh S, Branstrom AA, Colacino JM, Babiak J, Ju WD, Hirawat S, Northcutt VJ, Miller LL, Spatrick P, He F, Kawana M, Feng H, Jacobson A, Peltz SW, Sweeney HL. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007 May 3;447(7140):87-91.
- Youssoufian H, Antonarakis SE, Aronis S, Tsiftis G, Phillips DG, Kazazian HH Jr. Characterization of five partial deletions of the factor VIII gene. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3772-6.
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, Bulman DE, Harris JB, Davies KE. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990 Jan 11;343(6254):180-2.
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996 Aug 22;382(6593):722-5.
- Goulding C, McManus R, Murphy A, MacDonald G, Barrett S, Crowe J, Hegarty J, McKiernan S, Kelleher D. The CCR5-delta32 mutation: impact on disease outcome in individuals with hepatitis C infection from a single source. Gut. 2005 Aug;54(8):1157-61.
- Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C, Heap GA, Dubois PC, Clayton DG, Hunt KA, van Heel DA, Todd JA. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008 Dec 25;359(26):2767-77.
- Glass WG, McDermott DH, Lim JK, Lekhong S, Yu SF, Frank WA, Pape J, Cheshier RC, Murphy PM. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med. 2006 Jan 23;203(1):35-40.
- Wilton SD, Fall AM, Harding PL, McClorey G, Coleman C, Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol Ther. 2007 Jul;15(7):1288-96.
- Fletcher S, Adkin CF, Meloni P, Wong B, Muntoni F, Kole R, Fragall C, Greer K, Johnsen R, Wilton SD. Targeted exon skipping to address "leaky" mutations in the dystrophin gene. Mol Ther Nucleic Acids. 2012 Oct 16;1:e48.
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, van Ommen GJ, den Dunnen JT, van Deutekom JC. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am J Hum Genet. 2004 Jan;74(1):83-92.
- Nobile C, Marchi J, Nigro V, Roberts RG, Danieli GA. Exon-intron organization of the human dystrophin gene. Genomics. 1997 Oct 15;45(2):421-4.
- De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E, Cossu G, Bozzoni I. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48-50 DMD cells. Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9456-61.
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, Dickson G, Wood MJ, Wilton SD, Straub V, Kole R, Shrewsbury SB, Sewry C, Morgan JE, Bushby K, Muntoni F. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011 Aug 13;378(9791):595-605.
- Cirak S, Feng L, Anthony K, Arechavala-Gomeza V, Torelli S, Sewry C, Morgan JE, Muntoni F. Restoration of the dystrophin-associated glycoprotein complex after exon skipping therapy in Duchenne muscular dystrophy. Mol Ther. 2012 Feb;20(2):462-7.
- Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N, Holling T, Janson AA, Platenburg GJ, Sipkens JA, Sitsen JM, Aartsma-Rus A, van Ommen GJ, Buyse G, Darin N, Verschuuren JJ, Campion GV, de Kimpe SJ, van Deutekom JC. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med. 2011 Apr 21;364(16):1513-22.
- Koo T, Wood MJ. Clinical trials using antisense oligonucleotides in duchenne muscular dystrophy. Hum Gene Ther. 2013 May;24(5):479-88.
- Jain A, Ahmad F, Rajeswari MR. Structural studies on DNA triple helix formed by intronic GAA triplet repeat expansion in Friedreich's ataxia. Nucleosides Nucleotides Nucleic Acids. 2003 May-Aug;22(5-8):1517-9.
- Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497.
- de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982 Dec 23;300(5894):765-7.
- Swan DC, McBride OW, Robbins KC, Keithley DA, Reddy EP, Aaronson SA. Chromosomal mapping of the simian sarcoma virus onc gene analogue in human cells. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4691-5.
- Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985 Jun 13-19;315(6020):550-4.
- Maru Y. Molecular biology of chronic myeloid leukemia. Int J Hematol. 2001 Apr;73(3):308-22.
- Brown WM, Doll R. Mortality from cancer and other causes after radiotherapy for ankylosing spondylitis. Br Med J. 1965 Dec 4;2(5474):1327-32.
- Higashi Y, Yoshioka H, Yamane M, Gotoh O, Fujii-Kuriyama Y. Complete nucleotide sequence of two steroid 21-hydroxylase genes tandemly arranged in human chromosome: a pseudogene and a genuine gene. Proc Natl Acad Sci U S A. 1986 May;83(9):2841-5.
- Rodrigues NR, Dunham I, Yu CY, Carroll MC, Porter RR, Campbell RD. Molecular characterization of the HLA-linked steroid 21-hydroxylase B gene from an individual with congenital adrenal hyperplasia. EMBO J. 1987 Jun;6(6):1653-61.
- Higashi Y, Tanae A, Inoue H, Hiromasa T, Fujii-Kuriyama Y. Aberrant splicing and missense mutations cause steroid 21-hydroxylase [P-450(C21)] deficiency in humans: possible gene conversion products. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7486-90.
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003 May 8;423(6936):145-50.
- Higuchi S, Tsuboi M, Iitaka Y. Infrared spectrum of a DNA-RNA hybrid. Biopolymers. 1969 Jun;7(6):909-16.
- Whelan DR, Bambery KR, Heraud P, Tobin MJ, Diem M, McNaughton D, Wood BR. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acids Res. 2011 Jul;39(13):5439-48.
- Benham CJ. Theoretical analysis of transitions between B- and Z-conformations in torsionally stressed DNA. Nature. 1980 Aug 7;286(5773):637-8.
- Rich A, Nordheim A, Wang AH. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791-846.
- Brown TA. Chapter 1: The Human Genome. In: Genomes, 2nd edition. Oxford: Wiley-Liss; 2002.
- Wittig B, Dorbic T, Rich A. Transcription is associated with Z-DNA formation in metabolically active permeabilized mammalian cell nuclei. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2259-63.
- Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003 Jul;4(7):566-72.
- Zhabinskaya D, Benham CJ. Theoretical analysis of the stress induced B-Z transition in superhelical DNA. PLoS Comput Biol. 2011 Jan 20;7(1):e1001051.
- Lafer EM, Sousa R, Ali R, Rich A, Stollar BD. The effect of anti-Z-DNA antibodies on the B-DNA-Z-DNA equilibrium. J Biol Chem. 1986 May 15;261(14):6438-43.
- Boiteux S, Costa de Oliveira R, Laval J. The Escherichia coli O6-methylguanine-DNA methyltransferase does not repair promutagenic O6-methylguanine residues when present in Z-DNA. J Biol Chem. 1985 Jul 25;260(15):8711-5.
- Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci U S A. 2006 Feb 21;103(8):2677-82.
- Kha DT, Wang G, Natrajan N, Harrison L, Vasquez KM. Pathways for double-strand break repair in genetically unstable Z-DNA-forming sequences. J Mol Biol. 2010 May 14;398(4):471-80.
- Rich A. DNA comes in many forms. Gene. 1993 Dec 15;135(1-2):99-109.
- Buske FA, Mattick JS, Bailey TL. Potential in vivo roles of nucleic acid triple-helices. RNA Biol. 2011 May-Jun;8(3):427-39.
- Guillonneau F, Guieysse AL, Nocentini S, Giovannangeli C, Praseuth D. Psoralen interstrand cross-link repair is specifically altered by an adjacent triple-stranded structure. Nucleic Acids Res. 2004 Feb 13;32(3):1143-53.
- Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188-95.
- Lange SS, Vasquez KM. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol Carcinog. 2009;48:571-80.
- Bäckvall H, Asplund A, Gustafsson A, Sivertsson A, Lundeberg J, Ponten F. Genetic tumor archeology: mi-crodissection and genetic heterogeneity in squamous and basal cell carcinoma. Mutat Res. 2005 Apr 1;571(1-2):65-79.
- Ståhl PL, Stranneheim H, Asplund A, Berglund L, Pontén F, Lundeberg J. Sun-Induced Nonsynonymous p53 Mutations Are Extensively Accumulated and Tolerated in Normal Appearing Human Skin. J Invest Dermatol. 2011 Feb;131(2):504-8.
- Gilman A. The initial clinical trial of nitrogen mustard. Am J Surg. 1963 May;105:574-8.
- Arcamone F, Cassinelli G, Fantini G, Grein A, Orezzi P, Pol C, Spalla C. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Reprinted from Biotechnology and Bioengineering, Vol. XI, Issue 6, Pages 1101-1110 (1969). Biotechnol Bioeng. 2000 Mar 20;67(6):704-13
- Kufe DW, Pollock RE, Weichselbaum RR, et al., editors. Holland-Frei Cancer Medicine. 6th edition. Hamilton (ON): BC Decker; 2003.
- Takimoto CH, Calvo E. Principles of Oncologic Pharmacotherapy. In: Pazdur R, Wagman LD, Camphausen KA, Hoskins WJ (Eds) Cancer Management: A Multidisciplinary Approach. 11 ed. 2008.
- Crespi MD, Ivanier SE, Genovese J, Baldi A. Mitoxantrone affects topoisomerase activities in human breast cancer cells. Biochem Biophys Res Commun. 1986 Apr 29;136(2):521-8.
- Sinha BK, Katki AG, Batist G, Cowan KH, Myers CE. Differential formation of hydroxyl radical by adriamycin in sensitive and resistant MCF-7 human breast tumor cells: implication for the mechanism of action. Biochemistry. 1987;26:3776-81.
- Mazerski J, Martelli S, Borowski E. The geometry of intercalation complex of antitumor mitoxantrone and ametantrone with DNA: molecular dynamics simulations. Acta Biochim Pol. 1998;45(1):1-11.
- Fox EJ. Management of worsening multiple sclerosis with mitoxantrone: a review. Clin Ther. 2006 Apr;28(4):461-74.
- Marriott JJ, Miyasaki JM, Gronseth G, O'Connor PW. Evidence Report: The efficacy and safety of mitoxantrone (Novantrone) in the treatment of multiple sclerosis. Report of the Therapeutics and Technology Assessment Subcommittee of the American Academy of Neurology. Neurology. 2010 May 4;74(18):1463-70.
- Pascual AM, Téllez N, Boscá I, Mallada J, Belenguer A, Abellán I, Sempere AP, Fernández P, Magraner MJ, Coret F, Sanz MA, Montalbán X, Casanova B. Revision of the risk of secondary leukaemia after mitoxantrone in multiple sclerosis populations is required. Mult Scler. 2009 Nov;15(11):1303-10.
- Ellis R, Boggild M. Therapy-related acute leukaemia with Mitoxantrone: what is the risk and can we minimise it? Mult Scler. 2009 Apr;15(4):505-8.
- Auerbuch SD. Nonclassic alkylating agents. In: Chabner BA, Collins JM, editors. Cancer chemotherapy: principles and practice. Philadelphia: Lippincott; 1990. p. 314-28.
- Povirk LF, Shuker DE.DNA damage and mutagenesis induced by nitrogen mustards. Mutat Res. 1994 Dec;318(3):205-26.
- Dong Q, Barsky D, Colvin ME, Melius CF, Ludeman SM, Moravek JF, Colvin OM, Bigner DD, Modrich P, Friedman HS. A structural basis for a phosphoramide mustard-induced DNA interstrand cross-link at 5'-d(GAC). Proc Natl Acad Sci U S A. 1995 Dec 19;92(26):12170-4.
- Kang KY, Kim HO, Yoon HS, Lee J, Lee WC, Ko HJ, Ju JH, Cho CS, Kim HY, Park SH. Incidence of cancer among female patients with systemic lupus erythematosus in Korea. Clin Rheumatol. 2010 Apr;29(4):381-8.
- Khan MA, Travis LB, Lynch CF, Soini Y, Hruszkewycz AM, Delgado RM, Holowaty EJ, van Leeuwen FE, Glimelius B, Stovall M, Boice JD Jr, Tarone RE, Bennett WP. p53 mutations in cyclophosphamide-associated bladder cancer. Cancer Epidemiol Biomarkers Prev. 1998;7(5):397-403.
- Armand JP, Ribrag V, Harrousseau JL, Abrey L. Reappraisal of the use of procarbazine in the treatment of lymphomas and brain tumors. Ther Clin Risk Manag. 2007 Jun;3(2):213-24.
- Serrone L, Zeuli M, Sega FM, Cognetti F. Dacarbazine-based chemotherapy for metastatic melanoma: thirty-year experience overview. J Exp Clin Cancer Res. 2000 Mar;19(1):21-34.
- Rosenberg B; Van Camp L; Krigas T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature. 1965 Feb 13;205:698-9.
- Monneret C. Platinum anticancer drugs. From serendipity to rational design. Ann Pharm Fr. 2011 Nov;69(6):286-95.
- Jamieson ER, Lippard SJ. Structure, Recognition, and Processing of Cisplatin-DNA Adducts. Chem Rev. 1999 Sep 8;99(9):2467-98.
- Coluccia M, Natile G. Trans-platinum complexes in cancer therapy. Anticancer Agents Med Chem. 2007 Jan;7(1):111-23.
- Oliński R, Walter Z. Isolation of the adducts of platinum complexes and nucleic acid bases on the Dowex 50 W column. Z Naturforsch C. 1984 Nov-Dec;39(11-12):1052-6.
- Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005 Apr;4(4):307-20.
- Williams CJ, Whitehouse JM. Cis-platinum: a new anticancer agent. Br Med J. 1979 Jun 23;1(6179):1689-91.
- van der Vijgh WJ. Clinical pharmacokinetics of carboplatin. Clin Pharmacokinet. 1991 Oct;21(4):242-61.
- Raymond E, Chaney SG, Taamma A, Cvitkovic E. Oxaliplatin: a review of preclinical and clinical studies. Ann Oncol. 1998;9:1053-71.
- Tang CH, Parham C, Shocron E, McMahon G, Patel N. Picoplatin overcomes resistance to cell toxicity in small-cell lung cancer cells previously treated with cisplatin and carboplatin. Cancer Chemother Pharmacol. 2011 Jun;67(6):1389-400.
- Holford J, Sharp SY, Murrer BA, Abrams M, Kelland LR. In vitro circumvention of cisplatin resistance by the novel sterically hindered platinum complex AMD473. Br J Cancer. 1998;77(3):366-73.
- Martín M, Díaz-Rubio E, Casado A, Santabárbara P, López Vega JM, Adrover E, Lenaz L. Carboplatin. An active drug in metastatic breast cancer. J Clin Oncol. 1992;10:433-7.
- Sooriyaarachchi M, Narendran A, Gailer J. The effect of sodium thiosulfate on the metabolism of cis-platin in human plasma in vitro. Metallomics. 2012 Aug 24;4(9):960-7.
- Sooriyaarachchi M, Narendran A, Gailer J. N-acetyl-L-cysteine modulates the metabolism of cis-platin in human plasma in vitro. Metallomics. 2013 Mar;5(3):197-207.
- Lichtenstein AK, Pende D. Enhancement of natural killer cytotoxicity by cis-diamminedichloroplatinum (II) in vivo and in vitro. Cancer Res. 1986 Feb;46(2):639-44.
- Hato SV, de Vries IJ, Lesterhuis WJ. STATing the importance of immune modulation by platinum chemotherapeutics. Oncoimmunology. 2012 Mar 1;1(2):234-236.
- Kelland LR. Preclinical perspectives on platinum resistance. Drugs. 2000;59 Suppl 4:1-8; discussion 37-8.
- Stordal B, Davey M. Understanding cisplatin resistance using cellular models. IUBMB Life. 2007 Nov;59(11):696-9.
- Gargiulo D, Kumar GS, Musser SS, Tomasz M. Structural and function modification of DNA by mitomycin C. Mechanism of the DNA sequence specificity of mitomycins. Nucleic Acids Symp Ser. 1995;(34):169-70.
- Lockwood A, Brocchini S, Khaw PT. New developments in the pharmacological modulation of wound healing after glaucoma filtration surgery. Curr Opin Pharmacol. 2013 Feb;13(1):65-71.
- Maanen MJ, Smeets CJ, Beijnen JH. Chemistry, pharmacology and pharmacokinetics of N,N',N"-triethylenethiophosphoramide (ThioTEPA). Cancer Treat Rev. 2000 Aug;26(4):257-68.
- Chen MJ, Shimada T, Moulton AD, Cline A, Humphries RK, Maizel J, Nienhuis AW. The functional human dihydrofolate reductase gene. J Biol Chem. 1984 Mar 25;259(6):3933-43.
- Racanelli AC, Rothbart SB, Heyer CL, Moran RG. Therapeutics by cytotoxic metabolite accumulation: pemetrexed causes ZMP accumulation, AMPK activation, and mammalian target of rapamycin inhibition. Cancer Res. 2009 Jul 1;69(13):5467-74.
- Cerqueira NM, Fernandes PA, Ramos MJ. Understanding ribonucleotide reductase inactivation by gemcitabine. Chemistry. 2007;13(30):8507-15.
- Hawser S, Lociuro S, Islam K. Dihydrofolate reductase inhibitors as antibacterial agents. Biochem Pharmacol. 2006 Mar 30;71(7):941-8.
- Gombar VK, Guillonneau F, Guieysse AL, Nocentini S, Giovannangeli C, Praseuth D. Psoralen interstrand cross-link repair is specifically altered by an adjacent triple-stranded structure. Nucleic Acids Res. 2004 Feb 13;32(3):1143-53.
- Mayer J, Cheeseman MA, Twaroski ML. Structure-activity relationship analysis tools: validation and applicability in predicting carcinogens. Regul Toxicol Pharmacol. 2008 Feb;50(1):50-8.
- Povirk LF, Goldberg IH. A role of oxidative DNA sugar damage in mutagenesis by neocarzinostatin and bleomycin. Biochimie. 1987 Aug;69(8):815-23.
- Dedon PC. The chemical toxicology of 2-deoxyribose oxidation in DNA. Chem Res Toxicol. 2008 Jan;21(1):206-19.
- Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996 Aug 17;355(1-2):71-89.
- van der Burgt I, Chrzanowska KH, Smeets D, Weemaes C. Nijmegen breakage syndrome. J Med Genet. 1996;33(2):153-6.
- Allio T, Preston RJ. Increased sensitivity to chromatid aberration induction by bleomycin and neocarzinostatin results from alterations in a DNA damage response pathway. Mutat Res. 2000 Sep 20;453(1):5-15.
- Laczmanska I, Gil J, Karpinski P, Stembalska A, Trusewicz A, Pesz K, Ramsey D, Schlade-Bartusiak K, Blin N, Sasiadek MM. Polymorphism in nucleotide excision repair gene XPC correlates with bleomycin-induced chromosomal aberrations. Environ Mol Mutagen. 2007 Oct;48(8):666-71.