1. Emergence of the concept of normal variance in individual capacity for repair of DNA damage
Margaret Mead (1901–1978)
Genomic DNA is subjected daily to significant amounts of damage. In healthy individuals, the cellular machinery for repair generally manages the damage very efficiently, with only occasional lapses or errors. The latter may cause genetic and/or multifactorial disease (depending on whether it occurs on germline and/or somatic level), but allows for some changeability in the genome that constitutes one of the major drives in evolution. Even among clinically healthy individuals of the same species, however, the efficiency of protection against DNA damage (measured in terms of rate of removal of lesions in DNA or the proportion of unrepaired damage that persists after repair has been carried out) may vary. This heterogeneity may have diverse short-term and long-term effects – from difference in the rate of progression of phenotypic traits associated with ageing in individuals of the same sex and age (e.g. accelerated development of skin laxity and loss of subcutaneous fat, causing early occurrence of wrinkles in some individuals, etc.), to more serious consequences, such as earlier onset of senile cataract, increased risk of different types of cancer, differential response to damaging agents (e.g. during genotoxic treatments), etc. The degree of variance in individual capacity for repair of DNA damage is usually subtle, but may, in some cases, become significant, especially when modulated by other factors. The latter may be of endogenous origin (e.g. genetic factors) as well as exogenous (lifestyle and habits, environmental factors, etc.).
The concept of individual repair capacity (ICR) emerged in the late 70-ties of the XX century. It was first defined as the differences in the efficiency with which cultured cells taken from one individual repaired damage induced by controlled methods, compared to the efficiency of repair of the same type of damage in other individuals. Specifically, significant inter-individual variation was observed in the responses of cultured human lymphocytes taken from healthy volunteers to DNA damage induced by chemical agents [
Modern people live longer, and the active period of life continues several decades after the reproductive plans have been completed. It could be expected that the prevalence of cancer will continue to rise steadily. The modern biomedical science and the pharmaceutical industry develop rapidly, and more new therapies are being developed, providing if not a cure, then at least a considerable improvement in the quality of life for the patients. Nowadays, ICR rapidly becomes integral part of biomedical science. Assessments of individual repair capacity are currently used for research purposes, e.g. in studies of normal variance between healthy individuals and in human disease. It is also a major branch of the rapidly expanding field of individualised (personalised) medicine – tailoring of therapeutic approaches for the particular patient, prognostication of outcomes in terms of expected responses to therapy and possible adverse effects, etc. Knowledge about the specificities of individual capacity for detoxification of toxic metabolites and repair of DNA damage forms a base for making informed decisions, in normal everyday life as well as in treatment of disease. In healthy individuals it may become a basis for introduction of certain lifestyle changes, e.g. cessation of smoking, weight loss, increased consumption of fresh fruit and vegetables, wearing appropriate level of UV protection, etc. Personal knowledge about individual repair capacity may assist in the assessment of eligibility for specific therapies and the prognostication in terms of overall survival, uneventful survival, and the potential toxic effects of a particular therapeutic regimen.
At the moment, the largest body of experimental evidence about the role of individual repair capacity in human disease is in the field of assessment of risk for various cancers and potential outcomes of anticancer therapy. This is not unexpected, as proteins acting in DNA repair and maintenance of genome integrity normally work - directly or indirectly - to prevent uncontrolled cell proliferation. There are, however, numerous reports about the role of individual repair capacity in multifactorial disease other than cancer (diseases with inflammatory genesis, such as insulin resistance and atherosclerosis, cardiovascular disease, myelodysplastic syndromes, neurodegenerative disease, etc.) or in seemingly unrelated areas, such as human fertility and sterility. Panels of markers for assessment of ICR are currently developed and tested in clinically healthy patients and in selected patient groups in order to assess their applicability to assessment of risk for various diseases and conditions [
Analysis for individual capacity for repair of DNA damage and maintenance of genome integrity suffers, however, from some serious limitations. Theoretically, the information about the individual capacity for repair of DNA damage may be used for identification of individuals at risk for multifactorial diseases and conditions in which dysregulation of DNA repair and maintenance of genome integrity is known to play a role for the purposes of prophylaxis or early presymptomatic intervention. This, however, has but little value when performed in healthy individuals, as the test result may indicate increased risk for cancer, but cannot predict who of the individuals identified to be at risk would develop cancer at all, and if they do, at what age, and what the exact type of the tumour might be. After all, everyone is at risk for development of cancer, especially after the age of 50. Therefore, the knowledge about being at slightly elevated risk for cancer is likely to cause severe distress and anxiety without offering much alternative for informed action except for the regular check-ups that are recommended anyway.
Some of the polymorphic variants in DNA repair genes are associated with decreased risk for various diseases and conditions, that is, the risk for development of the associated disease or condition is lower in carriers of one or two copies of the variant allele compared to homozygous carriers of the wild type alleles (for examples, see below). This is usually not associated with increase in capacity for recognition and repair of DNA damage above the average in the carriers of variant alleles, but, rather, with the fact that the wild type allele is associated with mild (subclinical) repair deficiency.
Analysis of polymorphisms in DNA repair genes may, nevertheless, be used for identification of individuals at highest risk in a selected group of individuals that were found to be at risk for reasons other than genetic predisposition. For example, analysis of genetic factors predisposing to development of lung cancer (a tumour associated with tobacco smoking) in a cohort of smokers may be quite useful. Data about individual repair capacity may also be used in groups affected with specific disease or condition for identification of individuals at high risk for adverse events. For example, ICR status may aid in the assessment of risk for certain complications in a cohort of diabetic patients; in the identification of individuals at increased risk for a second vascular incident in a group of patients that have had a stroke already, etc. Finally, ICR may play a major role in the assessment of sensitivity of tumours to different anticancer therapies and susceptibility to therapy-associated toxicity.
The number of markers for assessment of capacity for DNA repair grows by the week, but not all of these markers offer reproducible results in vitro, and even less seem to be reliable enough in vivo. The reliability of the tests for individual repair capacity has been repeatedly questioned with regard to sample sizes, adequacy of controls and testing techniques[
The assessment of individual capacity for repair of DNA damage is implemented by using sets of markers, but it is much more than a simple sum of disparate markers. None of the markers described below has significant diagnostic or prognostic value on its own. At least several select markers (depending on the disease or the condition) must be analysed to obtain an informative and reliable result.
2. Individual repair capacity is not comprised solely of genetic polymorphisms
far more than our abilities.
J. K. Rowling, Harry Potter and the Chamber of Secrets (1998).
The capacity for repair of DNA damage is studied today on two major levels. The first level is research dedicated to genetic predisposition –that is, identification of polymorphic variants of genes functioning in DNA repair, damage-associated signalling, and maintenance of genome integrity and testing their reliability as markers. The second level uses the knowledge obtained at the previous level, integrating the data provided by analysis of the markers of individual status into the real phenotype of the individual. Thus, the efficiency of repair of cell damage is projected on a larger scale (tissues, organs and organism) in the context of the interplay between the genetic predisposition and the environment. Thus, the information stored in the genetic background of the individual (what it might have been) is analysed from the viewpoint of their present status (what really is). Reliable prognostication of risks for development of disease and/or the possible adverse outcomes may only be done on the higher level of research of individual repair capacity, as the disparate markers do not have much predictive value on their own. Indeed, having a genetic predisposition for a disease or condition does not mean that it would develop with 100% certainty. Even among carriers of mutations usually associated with development of severe monogenic disease there are occasional asymptomatic cases. For example, in the majority of cases, carriership of mutations in the MECP2 gene, coding for a protein that recognises methylated bases in DNA results in the phenotype of Rett syndrome in females, but there have been occasional reports about asymptomatic or mildly symptomatic carriers of mutations that normally cause a severe phenotype, usually because of skewed X-inactivation [
The specific effects of a DNA polymorphism on the phenotype may only be valid for some populations. For example, the 399Gln variant of the XRCC1 polymorphism (for details, see below) was found to be associated with a reduction of the risk for development of colorectal adenomas at high risk for conversion to carcinoma in the Norwegian population [
While genetic predisposition is important, it is rarely a critical factor and the information about the genetic background of the individual typically does not provide information about their present status. Phenotypic markers are the functional expression of the communication between the main predisposing gene/s, the associated modulating genetic factors, be it at gene, transcript or protein level, and the exogenous factors, and they reflect the current state of the tissue, the organ or the organism. The latter may reveal how the genetic background and the environmental factors intermingle to create a unified phenotype and may allow for more reliable decision-making and prognostication.
The overall capacity for repair in real time (at the moment of testing) is commonly measured by one of the methods described in the next chapter – namely, the unscheduled (replication-unrelated) synthesis of DNA. It may also be considered a major phenotypic marker, complementing and integrating the data obtained by other markers.
If we view the capacity for repair of damage in a broader sense, it implicitly includes the capacity for cell and tissue renewal. Indeed, as repair of DNA damage is a prerequisite to normal cell division, the capacity for repair and the capacity for tissue regeneration are tightly linked. As of now, telomere length, telomere attrition rate and/or telomerase activity –is successfully used as a phenotypic marker for assessment of potential of tissue regeneration. Usually, the telomere length and telomere attrition rate are reliable indicators for the cell's proximity to replicative senescence. The shorter the mean length of telomeres in a tissue, the closer the normal (non-transformed) cells of that tissue are to replicative senescence, therefore, the more limited the capacity for tissue renewal. Testing of the status for the genetic components of individual repair capacity is often complemented by testing for capacity for self-renewal in cells and tissues.
The markers for capacity for detoxification of harmful compounds and antioxidant capacity (usually, enzymatic activities) are still an important part of the panels for assessment for individual capacity for repair of DNA damage, although the associated enzymatic activities usually do not contribute directly to DNA repair. These enzymes generally work by degrading the toxic compound/s (e.g. drugs, but also other exogenous and endogenous agents), increasing or decreasing their solubility; modifying or conjugating them to other compounds so as to decrease their toxicity, etc., thereby modulating the amount of genotoxic stress in the cell. Thus, the capacity for detoxification of substances with genotoxic potential may be viewed as prerequisite for the normal functioning of the DNA repair machinery. The major markers for detoxification and antioxidant capacity that are currently in use are briefly described below.
3. Markers for detoxification and antioxidant capacity
Cytochrome Р450 superfamily
The proteins of the cytochrome Р450 (CYP) superfamily function in the metabolisation of many compounds associated with increased levels of oxidative stress in eukaryotic cells. Among these compounds are, for example, various types of drugs (antibiotics, NSAIDs; analgesics, e.g. tramadol; anticancer drugs like tamoxifen and cyclophosphamide; immunomodulators; antidiabetic drugs, e.g. sulphonylureas; anticoagulants such as warfarin and coumarin; and many others); hormones, ethyl alcohol; heterocyclic aromatic amines (e.g. produced during cooking, specifically frying); industrial chemicals – polycyclic aromatic hydrocarbons (e.g. in tobacco smoke and smoke from burning organic fuels); dioxin and dioxin-like compounds; and many others. Many variant alleles of the different isoenzymes of the CYP family have been identified, encoding proteins with relatively decreased enzymatic activity [
Glutathione S-transferase
Glutathione S-transferase (GST) is an umbrella term describing several tissue-specific enzymatic activities catalysing the conjugation of reduced glutathione to different electrophilic compounds (usually, with cytotoxic and genotoxic properties). Among the latter are compounds of exogenous origin (anticancer drugs–melphalan, chlorambucil), industrial chemicals (e.g. polycyclic aromatic hydrocarbons, halogenated hydrocarbons) as well as endogenous compounds (steroid hormones, prostaglandins, unsaturated aldehydes produced by oxidation of lipids, and others) [reviewed in
Null alleles for the genes coding for two of the GST isoenzymes, GSTT1-1 and GSTM1-1 may be observed in a significant proportion of people in virtually all human populations, although there are ethnic differences in the prevalence of the deletion allele [
One C-to-T transition in the GSTT1 gene (rs4630) may be associated with lower incidence of therapy-induced peripheral neuropathy in patients with multiple myeloma treated with thalidomide [
The expression of some of the isoforms of GST (specifically, GSTP1-1) may be selectively up-regulated in tumours, providing resistance to anticancer drugs [
Methylene tetrahydrofolate reductase
Methylene tetrahydrofolate reductase (MTHFR) is a key enzyme of the folate metabolism catalysing the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate in the biosynthesis of methionine from homocysteine. Two common polymorphisms associated with significantly decreased enzymatic activity have been described in the MTHFR gene– the already mentioned C677T (Ala222Val, rs1801133) and A1298C (Glu429Ala, rs1801131) [reviewed in
In patients homozygous for the T allele of C677T and the heterozygous genotype of A1298C, severe toxicity effects may occur during or after therapy with antimetabolite agents. Carriers of the TT genotype by the C677T polymorphism of the MTHFR gene and/or at least one A allele of the MTHFR 1298A>C polymorphism treated for acute leukemia with fluoropyrimidines are at increased risk for haematological and gastrointestinal toxicity [
Superoxide dismutases
Superoxide dismutases (SODs) catalyse the conversion of superoxide radicals to molecular oxygen and hydrogen peroxide. Impaired function of SODs results in increased levels of reactive oxygen species in the cell. In mammalian cells there are three basic types of SODs: cytosolic copper-zinc (SOD1); mitochondrial manganese (MnSOD, SOD2), and extracellular copper-zinc SOD (SOD3).
The Ala16 variant of the SOD2 Ala16Val polymorphism (rs4880) is associated with lower enzymatic activity than the Val16 variant allele [
The Gly allele of the Arg213Gly (rs8192291) polymorphism in the SOD3 gene is associated with significant (over 10-fold) increase in enzymatic activity [
Carriership of the SOD2 Ala16Val polymorphism and the SOD3 Arg213Gly polymorphism may be associated with predisposition to obstructive lung disease, parenchymal lung diseases (idiopathic pulmonary fibrosis and lung granulomatosis) and lung malignancies[
As diabetes is a disease associated with increased oxidative stress, polymorphisms in SOD genes may modulate the risk for development of diabetes and some of its late complications. Specifically, theAla40Thr polymorphism in the gene coding for SOD3 may be implicated in the pathogenesis of insulin resistance and diabetes type 2, with carriers of the Thr40 allele exhibiting lower insulin sensitivity and earlier age at diagnosis than carriers of the Ala40 allele [
The SOD3 Ala40Thr polymorphism is suspected to be implicated in the risk for pre-eclampsia and severe foetal growth restriction [
SOD2 codon 16 heterozygous Ala/Val genotype is associated with increased risk of acute toxicity and subcutaneous fibrosis after radiotherapy compared to the Val/Val genotype [
Somatic mutations in genes coding for superoxide dismutases may occur in the course of neoplastic transformation [
Thiopurine S-methyltransferase (TPMT)
Thiopurine methyltransferase methylates thiopurine compounds, using S-adenosyl-L-methionine as donor of methyl groups. TPMT catalyzes the S-methylation of chemotherapeutics such as 6-mercaptopurine, 6-thioguanine and azathioprine [
Dihydropyrimidine dehydrogenase (DPYD)
Dihydropyrimidine dehydrogenase is the initial and rate-limiting enzyme in the catabolism of the pyrimidine bases uracil and thymine. It is the key enzyme catalysing the metabolic degradation of 5-fluorouracil (5-FU), with more than 85% of the administered 5-FU metabolised by DPYD [reviewed in
Uridine diphosphate glucuronosyltransferase (UGT1A1)
The UDP-glucuronosyltransferase family of enzymes catalyzes the transfer of a glycosyl group from UDP-glucuronate to an acceptor molecule [
4. Markers for capacity for DNA repair and maintenance of genome integrity
4.1. Markers for capacity for repair of DNA damage
At present, polymorphisms in over two dozen genes coding for major proteins of DNA repair and maintenance of genome integrity are considered to be reliably associated with significant effects on the phenotype in health and disease. These genes may be grouped by function as follows:
- Nucleotide excision repair: XPA, XPD, XPG (ERCC5), XPC and RAD23B (the latter two – only in global genome repair), ERCC1;
- Base excision repair: hOGG1, POLB, XRCC1, APEX1;
- Repair of strand breaks: XRCC2, XRCC3, NBS1(functioning in repair by homologous recombination); LIG4 and XRCC4 (repair by non-homologous end joining).
- Mismatch repair: MLH1, MSH6, EXO1;
- Maintenance of genome integrity/induction of damage-related cell cycle arrest and/or apoptosis: TP53, ATM, BRCA1 and BRCA2, RFC1, PARP1 gene and PARP1 pseudogene;
- Progression through cell cycle – Cyclin H (CCNH) gene.
In most cases, carriership of polymorphisms in these genes affects the susceptibility for development of different cancers or other multifactorial conditions. The impact on the outcomes of genotoxic therapies and the risk for development of drug resistance, however, has been studied thoroughly only for some of these polymorphisms. Several panels of markers have been tested so far, in patients with cancer as well as immunological diseases, with variable success [
4.1.1. Polymorphisms in NER genes
XPA gene
The XPA is part of the XPA-RPA complex that binds to damaged DNA in the early stages of repair by nucleotide excision and stimulates the endonuclease activity of XPF and XPG. Several relatively common single-nucleotide polymorphisms were described in the XPA gene, for two of which, (Arg228Gln (rs1805160) and Val234Leu (rs3176749)) no significant differences in the DNA repair capacity or survival of cells carrying the one or the other allele were found [
XPC gene
XPC, as a component of the XPC-hHR23 complex, is among the first molecules arriving at the sites of DNA damage in the non-coding regions of the genome. Its binding to the damage site serves as a signal for recruitment of the other factors of NER in order to initiate repair of the lesion. Several polymorphisms have been identified in the XPC gene. Among these, the best studied is the XPC ins83 polymorphism, an insertion/deletion of a 83 bp long poly-AT region in intron 9 of the gene coupled with a 5bp deletion, also in intron 9[
Polymorphisms in the XPC gene were shown to modulate the levels of unrepaired damage in human cells treated with ionising radiation and other genotoxic agents in in vitro settings [
Carriership of the XPC ins83 polymorphism may be associated with increased risk for diseases other than cancer. In a study from 2014, homozygous carriership of the deletion allele (repair-proficient) was significantly more common in age-matched healthy controls than in a cohort of patients with one cerebrovascular incident[
RAD23B gene
RAD23B is the partnering protein of XPC in the recognition of damage reparable by NER in the non-coding regions of the genome. The polymorphismAla249Val (rs1805329) in the RAD23B gene is associated with increased risk for breast cancer, either alone or with combination with polymorphisms in the XPC gene [
XPD gene
XPD is one of the two basic DNA helicases unwinding DNA at damage sites in order to allow unobstructed access of the NER repair machinery. Several polymorphisms in the XPD gene are associated with increased risk for carcinoma of the prostate gland, the breast and the lung and head and neck cancers. Among these are Arg156Arg (rs238406, a synonymous substitution of C with A), Asp312Asn (rsl799793) and Lys751Gln (rs1052559)[
The carriership of the polymorphismsAsp312Asn and Lys751Gln is associated with significant increase of the risk for development of senile cataract resulting from accelerated ageing of the crystallin proteins in the lens because of accumulation of unrepaired UV-induced damage [
ERCC1 gene
The ERCC1-XPF complex functions at the late stages of NER, introducing the 5'-strand break in the repaired strand. Several polymorphisms in the ERCC1 gene have been described. The C8092A polymorphism (rs3212986) has no coding value, as it is in the 3'-untranscribed region of the gene. The C allele, however, is associated with lower transcript stability [
XPG (ERCC5) gene
XPG (ERCC5) is a protein with endonuclease activity acting at the late stages of NER, cleaving the repaired strand in the 3'-direction from the damage site. Several polymorphisms in the ERCC5 gene have been described, but only the synonymous His46His (rs1047768) and His1104Asp substitutions (rs17655) were found to be associated with effects on the phenotype. Carriership of the Asp variant of the His1104Asp polymorphism may be associated with decreased risk for cancer of the pharynx, the oesophagus, and the lung [
4.1.2. Polymorphisms in BER genes
NEIL genes
It was already mentioned that until several years ago inherited deficiency of DNA repair was believed to result in such severe phenotypes that the affected foetuses were very rarely carried to term and/or died in the early postnatal period. Shortly after the discovery that carriership of mutant variants of NEIL1 gene in mice resulted in a phenotype similar to the human metabolic syndrome [
hOGG1 and POLB genes
Variant alleles of two genes coding for products with roles in base excision repair have been found to modulate the risk for bladder cancer [
The Cys/Cys genotype of the hOGG1 Ser326Cys polymorphism (rs1052133) is associated with decreased risk for prostate cancer [
Inherited defects in the hOGG1 gene (3p25) were found to be associated with predisposition to familial and sporadic renal clear cell carcinoma [
Children carriers of Ser/Ser or Ser/Cys genotypes by the Ser326Cys polymorphism of hOGG1 were identified to be at decreased risk for development of acute childhood lymphoblastic leukemia [
Carriership of polymorphisms in BER genes is associated with susceptibility to Grave's disease, a toxic hyperfunction of the thyroid gland that affects virtually all organs and systems. The etiology of Grave's disease is believed to be primarily autoimmune. The role of accumulation of oxidative DNA damage, however, was unequivocally confirmed, specifically for Grave's ophtalmopathy [
XRCC1 gene
The XRCC1 protein is a stabilising factor of the primary ligase of base excision repair, DNA ligase III. It is also a component of the single-strand break repair complex together with ligase III, PNK and DNA polymerase beta. Over 20 non-synonymous polymorphisms have been described in the XRCC1 gene. Of these, association with predisposition to multifactorial disease has been found for the non-synonymous substitutions Arg194Trp (rs1799782), Arg399Gln (rs25487), Arg280His (rs25489) and His107Arg (rs2228487) [
The His280 allele of the XRCC1 Arg280His polymorphism is associated with increased risk for development of colorectal adenoma in the Norwegian population [
Carriership of the Trp194 variant was found to be associated with early-onset colorectal carcinoma in the Egyptian population [
The effects of carriership of the Arg399Gln polymorphism on the risk of colorectal carcinoma have been reported to be different in different populations [
Heterozygous carriership of the Arg399Gln polymorphism the XRCC1 gene, in combination with the homozygous Asn/Asn genotype by the XPD Asp312Asn polymorphism is a genetic factor associated with increased risk for senile cataract [
APEX1 gene
The apurine/apyrimidine endonuclease APE1 hydrolyses the phosphodiester bond at the 5'-side of abasic sites generated in the first phases of BER, producing a single-strand break. The Asp148Glu polymorphism (rs1130409) is quite common (35–40% of all alleles) and reportedly has no detectable impact on the endonuclease and DNA binding activities of APE1 protein [
The G allele-containing genotypes of the polymorphism-656T>G in the promoter of the APE1 gene are associated with decreased risk for cervical cancer and breast cancer[
Association between polymorphisms in genes of BER and outcomes after transplantations of haematopoietic cells
Transplantation of haematopoietic stem cells is currently one of the best treatment options for patients with haematological cancers, aplastic anemia and various myelodysplastic states. Usually, the dysfunctional bone marrow of the recipient of the transplanted cells is ablated before they receive the transplant, as part of the transplantation protocol, in order to minimise the risk of subsequent proliferation of a dysplastic haematopoietic clone originating from residual cancer cells. The myeloablative therapy (conditioning) used to destroy the recipient's haematopoiesis before the transplantation works by induction of enough DNA damage to kill the rapidly proliferating cancer cells. Conditioning agents induce DNA damage mainly by base modification (alkylation and oxidation), introduction of strand breaks (e.g. topoisomerase inhibitors) and DNA crosslinks. Some of the drugs typically used in conditioning regimens are busulfan, cyclophosphamide, thioTEPA, cytosine arabinoside, etoposide and others. For the purposes of myeloablation, these agents are often used in larger dosages than in therapy of solid tumours (where myelosuppression is an adverse effect) and may sometimes be combined with radiation therapy, so as to eradicate 'stores' of cancer cells protected from chemotherapy.
The individual capacity for repair of DNA damage may be important variables when assessing the chances of success of transplantations of allogeneic haematopoietic stem cells in myeloablated recipients in terms of post-transplantation survival and transplant-related mortality; the risk for resistance to conditioning therapy and/or associated toxicity. Several polymorphisms in genes coding for proteins with roles in BER have been described that are associated with the risk for one-year transplant-related mortality – namely, in the TDG gene(G/T thymidine glycosylase, SNPs 166+529G>A (rs167715) in intron 2 and 23+789A>T (rs2374327) in intron 1); hOGG1 gene(SNP 842T>C (rs159153) in the 5'-region of the gene);and in MUTYH (A/G-specific adenine DNA glycosylase, SNPs -506G>A (rs3219463) and 8473T>G (rs3219476), both in the non-coding region of the gene) [
4.1.3. Polymorphisms in genes coding for proteins functioning in strand break repair
XRCC2 and XRCC3 genes
XRCC2 and XRCC3 are members of the RAD51 gene family, coding for proteins involved in repair by recombination. Homozygous carriers of the variant allele of any of the three polymorphisms in the 5'-noncoding UTR of the XRCC2 gene (rs10234749 (C>A), rs6464268 (T>C) and rs3218373 (G>T)) and the Arg188His (rs3218536) polymorphism in exon 3are at reduced risk for bladder cancer compared to carriers of the more common allele [
The Arg188His polymorphism and the Thr241Met (rs861539)polymorphism in the XRCC3 gene have recently been identified as associated with increased risk for head and neck cancers[
XRCC3 Thr241Met polymorphism is weakly associated with risk for breast cancer [
NBS1 gene
NBS1 protein (NBN, nibrin) is a component of the MRN (MRE11-RAD50-NBS1) complex recognising and processing free DNA in double-strand break repair. One rather uncommon polymorphism (Ile171Val (rs61754966), prevalence of about 1% in the general population) may be associated with predisposition to aplastic anemia and acute lymphoblastic leukemia [
LIG4 gene
Ligase IV is responsible for ligation of free ends in repair of double-strand breaks by non-homologous end joining (NHEJ) and the V(D)J recombination in immunocompetent cells. Several polymorphisms in the LIG4 gene – namely, the Thr9Ile (rs1805388) polymorphism, the Ile658Val (rs2232641) and the synonymous Asp568Asp polymorphism (rs1805386) were recently found to be implicated in the susceptibility to cancers of head and neck [
XRCC4 gene
XRCC4 is an auxiliary factor of Ligase IV, enhancing its end-joining activity. Carriership of the Ser allele of the Ala247Ser polymorphism (rs373409) is associated with lower levels of expression of the XRCC4 gene [
4.1.4. Polymorphisms in mismatch repair genes
MLH1 gene
MLH1 is a human MutL homologue, functioning in the early phases of mismatch repair. One polymorphism in the promoter of the gene, MLH1 -93 G>A was found to be associated with increased risk for colon cancer [
Several substitutions of non-conserved amino acid residues were described in the MLH1 gene. Among these are the Val219Ile(rs1799977) and Ser406Asn (1217G>A)polymorphisms that were also initially believed to be deleterious mutations associated with HNPCC but were later classified as neutral substitutions, not associated with significant increases in risk for cancer [
MSH6 gene
Human MSH6 protein functions in the recognition of single-base mismatches in DNA and is one of the many components of the BRCA1-associated genome surveillance complex (BASC), involved in recognition and repair of structural damage in DNA. Homozygous carriership of the variant allele of the MSH6 polymorphism Gly39Glu (rs1042821) is associated with increased risk of colon [
EXO1 gene
Exonuclease 1 is a 5'-3' DNA exonuclease excising DNA fragments containing mismatched nucleotides. Multiple polymorphisms in the EXO1 gene have been described, in the non-coding regions as well as in exons of the gene. One polymorphism, EXO1 Glu589Lys (rs1047840) was found to be associated with susceptibility to lung cancer, breast cancer and gastric cancer in Asian populations [
One of the non-coding polymorphisms in the EXO1 gene –rs3902093 (C>A, in the promoter region)is associated with reduced risk for melanoma [
The Leu allele of another EXO1 polymorphism (Leu757Pro, rs9350) may also be associated with decreased risk for colorectal cancer [
Sometimes, carriership of polymorphism/s in the genes coding for products functioning in DNA repair may affect other gene/s coding for products with roles in repair and/or maintenance of genome integrity. For example, carriership of the variant alleles of the XPD Asp312Asn and the XRCC1 Arg399Gln polymorphisms was found to be associated with higher rate of occurrence of TP53 mutations in non-small cell lung cancer [
The risk for occurrence of additional mutations may be modified by exogenous factors. For example, carriership of the Ser allele of the Ala247Ser polymorphism in the XRCC4 gene is associated with increased risk for introduction of mutations in the TP53 gene in individuals exposed to aflatoxin B1, but the risk is higher in individuals that have had more exposition to the harmful agent [
4.2. Polymorphisms in genes coding for products functioning in the maintenance of genome integrity
4.2.1. TP53 gene
The TP53 gene codes for the protein p53, a major tumour suppressor gene with roles in the maintenance of genomic integrity, induction of damage-associated cell cycle arrest, programmed cell death, and other critically important processes in eukaryotic cells. Over 200 naturally occurring polymorphic variants have been described so far in the TP53 gene, of which 19 are in the coding sequence of the gene [
The duplication allele of the 16 bp duplication polymorphism (A2) was found to be associated with lower levels of p53 mRNA and, respectively, with less efficient damage-associated cell cycle arrest, DNA repair and induction of apoptosis in cultured cells [
The studies on the Pro72Arg polymorphism are developed in much more detail than any other single-nucleotide substitution in the TP53 gene. This is, on the one hand, due to the fact that it was one of the first polymorphisms in crucially important genes that had been shown to have no immediate deleterious effects on the phenotype, and, on the other hand, because of the profound and multifaceted consequences that carriership of the one or the other form might have in healthy people and in individuals with specific diseases and conditions; and in young and advanced age. As was already mentioned, the Pro72 and Arg72 variants of the TP53 gene exhibit identical conformations, DNA-binding affinities and capacities for transactivation of target proteins; and are both essentially considered wildtype[
Pro72 is believed to be the ancestral allele of the TP53 gene, with the Arg72 allele arising as a result of a single-nucleotide substitution that had occurred about 40,000 years ago [
Role of p53 variants in cancer, age-related disease and longevity
As p53 regulates the checkpoints and pathways restricting uncontrolled proliferation, it could be expected that the risk for development of cancer may be influenced by carriership of the different variant alleles of the Pro72Arg polymorphism. One may assume that carriership of the Arg/Arg genotype would be associated with decreased risk for development of cancer because of increased chance for removal of damaged cells before they turn cancerous. Indeed, this may be observed sometimes in hereditary cancer syndromes as well as for cancers of somatic origin. For example, the age of onset of the first symptomatic tumour in HNPCC was found to be several years later in patients with the Arg/Arg genotype of the Pro72Arg polymorphism than in carriers of at least one Pro allele, especially the Pro/Pro homozygotes [
The Pro72Arg polymorphism may act in a synergistic manner with other polymorphisms to decrease risk for cancer. Recently, it has been shown that the combined carriership of the Arg72 allele and the TT genotype for the rs4938723 T>C polymorphism in the promoter of the gene coding for miRNA34C, a microRNA functioning in post-transcriptional silencing, may be associated with significantly decreased risk for colorectal carcinoma[
Association of carriership of the pro-apoptotic allele of the Pro72Arg polymorphism with good outcome is not, however, universally valid for all cancers and in all populations, and may be modified by additional factors. For example, co-carriership of the Arg allele together with the CC genotype of the same rs4938723 polymorphism in the miRNA34C gene is associated with greatly increased risk (>10 times) for hepatocellular carcinoma in some Asian populations [
The effect of the carriership of the Pro72Arg polymorphism on the risk of cervical cancer may vary indifferent populations. In a large study from 2012,the overall risk for cervical cancer in Pro/Pro genotype carriers was shown to be 25% increased when compared to heterozygous or homozygous carriers of Arg72 in the Indian population, but not in the Chinese, Japanese and Korean populations [
The effects of carriership of the one or the other variant of Pro72Arg on susceptibility to cancer may be age-dependent. For example, the proline variant of TP53 showed association with increased risk for some types of cancer (lung carcinoma) developing in old age (>60), while the arginine variant was more common in younger patients with the same type of cancer [
When cancer has already developed, carriership of the Arg alleles for the TP53 Pro72Arg polymorphism may be associated with unexpected effects. Studies of HPV-related cervical cancer show stark prevalence of Arg/Arg carriers in the cancer-affected cohorts (between 64 and 73% in different studies and in different populations), but not in cohorts of patients with cervical intraepithelial neoplasia (CIN), which is a precancerous state to cervical carcinoma [
When apoptosis is a major factor in the pathogenesis of human disease, carriership of the Arg72 allele may be associated with poorer outcomes. For example, insulin resistance is essentially an inflammatory state associated with increased levels of oxidative cell damage [
An association between the risk for development of some of the complications of diabetes and the carriership of different variants of TP53 has also been suspected. At present, development of complications in diabetes is largely unpredictable, as it may be not directly related to glucose levels. Diabetic neuropathy, accelerated vascular ageing and renal injury are commonly seen in diabetes type 2, but not in all patients. Presumably, increased propensity for apoptosis may be predispose the diabetic patient to tissue damage secondary to increased levels of oxidative damage. As of now, however, no significant associations between the carriership of the one or the other from of the Pro72Arg polymorphism and complications of diabetes have been identified [
Mass cell apoptosis is a major event in the development of atherosclerotic plaque. The latter is believed to enhance the formation of thrombi [
Functional outcome after stroke is largely unpredictable, apparently depending on factors that cannot be assessed at the time of admission. Research shows, however, that the Arg/Arg TP53 genotype may confer vulnerability of neurons to apoptosis, predicting poorer functional outcomes and higher risk of development of neurological decline after stroke [
Role of carriership of the TP53 Pro72Arg variant for fertility in mammals
p53-related pathways may play a significant role in human fertility[
p53-dependent pathways function in oocyte development and oocyte selection in mice [
A relationship between the p53 Pro72Arg polymorphism and the reproductive success in humans has also been elicited. The effect is, however, age-dependent. The reproductive disadvantage conferred by the carriership of the Pro72 allele was significant only in primiparas older than 30 years of age at the time of birth of the child [
4.2.2. ATM gene
As the ATM gene is highly conserved, mutations in the gene usually produce the phenotype of 'classic' ataxia-telangiectasia (A-T) or the milder A-T variant when in homozygous or compound heterozygous. Heterozygous carriers of defective ATM alleles, however, although generally asymptomatic, have been found to be at increased risk of death at any age due to any causes (apart from accidents and natural disasters, of course), including death from cancer and ischemic heart disease [
One polymorphism that does not disrupt the function of the ATM protein (the rs189037 G>A substitution in the promoter of the gene) was found to be associated with idiopathic non-obstructive azoospermia [
4.2.3. BRCA1 and BRCA2 genes
These two genes code for proteins functioning together as a complex in DNA damage-associated signalling. Several polymorphic variants in these two genes have been already described. Association with increased risk for cancer has so far been found for the coding polymorphisms BRCA1 Gln356Arg (rs1799950), Pro871Leu (rs799917)and Ser1512Ile; and BRCA2 Asn372His (rs144848) and Lys1132Lys (3624A/G, rs1801406). These polymorphisms are, predictably, associated predominantly with increased risk for breast cancer [
Carriership of the His/His homozygous genotype by the BRCA2 Asn372His polymorphism has been found to be modulate foetal survival, probably in a sex-dependent manner (favouring male sex) [
Several other polymorphisms in the BRCA1 gene (rs8176318 (G>T, in the 3'-untranslated region of the gene), rs1060915 (C>T, synonymous Ser1389Ser substitution) and rs16940 (C>T, synonymous Leu724Leu), rs206115 (-1371C>T) and rs206117 (-1135C>T)have been found in association with increased risk for contralateral cancer in female carriers of BRCA1 mutations [
4.2.4. RFC1 gene
Replication factor-1 (RFC-1) is one of the five subunits of the RFC complex, functioning as an accessory factor to eukaryotic DNA polymerases δ and ε during replication; and a component of the BRCA1-associated genome surveillance complex (BASC). One polymorphism in intron 1 of the gene coding for RFC-1 (rs6844176) is associated with high-grade acute graft-versus-host disease (GVHD) in recipients of allogeneic haematopoietic stem cells [
4.2.5. PARP1 gene and PARP1 pseudogene
Poly-(ADP-ribose)-polymerases family member 1 (PARP1) is a nuclear protein functioning as a primary damage sensor in the presence of DNA breaks, recruiting the DNA repair machinery to the damage site. Carriers of the variant allele of the Val762Ala polymorphism (rs1136410) in the PARP1 gene are at increased risk for bladder cancer compared to individuals homozygous for the common allele[
Development of chronic GVHD in patients with transplantations of allogeneic haematopoietic cells for treatment of leukemia may be associated with longer post-transplantation survival [
A processed pseudogene of the PARP1 gene may be present on the 13q chromosome arm in some individuals (but not in all). Chromosomes lacking the pseudogene copy (deletion allele)are seen with frequency about 3 times higher in Blacks than in Caucasians[
4.2.6. CCNH gene
Cyclin H (CCNH) is one of the components of cyclin-activating kinase (CAK), which is a component of the transcription factor TFIIH. The polymorphism Val270Ala (rs2266690) is associated with elevated risk for chronic lymphocytic leukemia [
4.3. Individual capacity for repair of experimentally induced double-strand breaks
The individual capacity for repair of experimentally induced DSBs and chromatid breakage (mutagen challenge assay) has been under intensive study as a potential marker of individual repair capacity, specifically in the identification of individuals at elevated risk for development of various cancers [reviewed in
Carriership of some of the polymorphisms in genes coding for proteins of DNA repair and maintenance of genome integrity may cause increased genomic instability. Specifically, the polymorphisms Lys939Gln in the XPC gene, Lys751Gln in the XPD gene, Asp1104His in the XPG gene, Arg399Gln in the XRCC1 gene and Thr241 Met in the XRCC3 gene were found to be associated with increased levels of strand breaks and chromosomal aberrations in peripheral lymphocytes from clinically healthy human volunteers [
4.4. Markers for capacity for cell and tissue renewal – telomere length, telomere attrition rate and telomerase activity
Replication of DNA proceeds in the 5'- to 3'- direction by adding deoxynucleoside phosphates to the 3'-hydroxyl end of the growing polynucleotide chain. Most DNA polymerases cannot initiate synthesis on a purely single-stranded template, but need a partially double-stranded region providing the free 3'- end. The inherent directionality of the DNA synthesis means that the 3'-5' (leading) strand can be replicated continuously to the very end, provided that the template is undamaged and that all deoxynucleoside triphosphates are readily available. The replication of the 5'-3' (lagging) strand, however, is carried out in a discontinuous fashion, in separate fragments that are synthesised in a direction opposite to the general direction of the growth of the strand and subsequently ligated to one another. Eventually, on the lagging strand, the DNA polymerase would reach a point in which it cannot add any more nucleotides as the template has reached its end and the primer has nowhere to attach (Fig. 1). This end of the template would, therefore, remain 'unfinished' (ending in a single-strand tail).
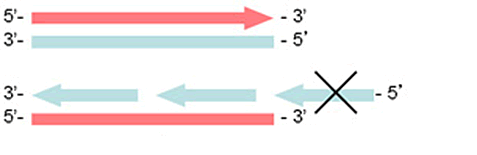
Every subsequent cell division would cause further shortening, leading to progressive loss of genetic information. Indeed, 50–120 bp of DNA are lost from ends of eukaryotic chromosomes per division cycle. The free ends may fuse together in a more or less random fashion, resulting in genomic rearrangements. To avoid loss of coding DNA sequence from the subterminal chromosome regions and generation of free reactive DNA ends, linear chromosomes in eukaryotic cells are protected by specific nucleoprotein complexes called telomeres. The telomeric DNA of higher eukaryotes consists of tandemly repeated modules made of 6 or 8 nucleotides (in mammals, including humans, the repeated unit is usually a hexanucleotide). In man, the sequence of the repeated unit of telomeres is TTAGGG. The length of the repeat may vary greatly, from 300–400 bp in yeast to >10 kb in man [
In some types of cells the DNA that was lost during replication may be restored by resynthesis of the repeated DNA from the chromosome ends. This is function of the telomere complex. It possesses its own integral nucleic acid component, providing the free 3'-OH ends needed for priming of the DNA synthesis (telomerase RNA component, TERC) as well as a catalytic subunit (telomerase reverse transcriptase, TERT) [
In normal (non-cancerous) cells replicative senescence usually begins when the telomere length shortens beyond a certain critical length. Onset of replicative senescence is not, however, a one-off decision, triggered by the first occurrence of chromosomes with shortened telomeres in the cell. It has been demonstrated that telomeres with length shorter the critical length may have been accumulating in the cell for quite some time before replicative senescence actually began [
Childhood adversities, acute or chronic stress in adults and even seemingly unrelated factors such as work schedule may reflect on telomere length and attrition rate in human cells [
There are heritable polymorphisms in the TERT and TERC gene that may modulate the telomere length and/or the rate of telomere attrition, but these are rare, and the rates of attrition of chromosome ends very rarely tally with prognoses made purely on the basis of genetic background. Thus, assessments of telomere length, telomere attrition rate or, in some tissues, telomerase activity are now increasingly used as a monitoring tool in biomedical research as well as in clinical medicine. They are usually added to the panel of genetic markers for assessment of individual repair capacity as a phenotypic marker of the capacity for cell and tissue renewal.
It is currently believed that it measurement of the absolute length of telomeres is less reliable than the rate of telomere shortening, as telomere length may vary even in physiological settings. Therefore, repeated assessments of telomere length at fixed time intervals may be needed to increase the reliability of the test result. In tissues in which the telomerase activity is normally preserved, the latter may be measured as well.
5. The dark side of the individual capacity for repair – response to anticancer therapy
I pray you be courteous with me.
Attributed to St. Francis of Assisi (c. 1181 – 1226),
prior to the cauterisation of one of his eyes with a red-hot iron.
One may quite safely assume that higher efficiency of DNA repair would mean lower risk for neoplastic transformation, as the damage would presumably be repaired before it resulted in transmissible mutations in DNA that might, potentially, induce uncontrolled cell growth. Let us, however, consider the case when the cell has already become transformed. At present, the majority of treatment options are targeted at destruction or severe suppression of the growth of the mutant cell progeny. This is usually achieved using DNA-damaging agents – chemotherapeutic agents, ionising radiation or, sometimes, other types of electromagnetic. These treatments are intended to drastically increase the level of DNA damage in the tumour cells in order to induce damage-dependent programmed cell death (if the cells have not already abrogated the signalling pathways for induction of apoptosis) and/or inhibit their cycling so that the growth of the tumour would slow down, possibly becoming manageable by other treatments. As cancer cells divide rapidly, genotoxic treatments would presumably affect them more severely than the normal cells of the tissue, although normal tissues would suffer too (see below). Tumour cells with lower-than-normal capacity for repair would rapidly accumulate suprathreshold levels of therapy-induced damage, causing them to slow down or stop the progression in the cell cycle or reroute to apoptosis. If the repair capacity of the tumour cell is preserved or is higher than normal, however, it may repair the damage produced by genotoxic treatments quickly and efficiently or with only a minor delay; and would continue with cell division. The latter may occur because the cell carries variant alleles of genetic polymorphisms conferring high capacity for repair of DNA damage or because it has had, as some point, acquired de novo the capacity to repair damage rapidly (e.g. because of a somatic mutations). Thus, high capacity for repair of DNA damage may be good for normal (non-transformed) cells, but might not be nearly that good when cancer has already developed, as it may render the tumour resistant to treatment, or treatments may require such high doses of genotoxic agent/s to be effective, that the toxic effects might outweigh its benefits [
Anticancer therapies based on genotoxic impact are very often associated with acute and/or late toxic effects. Cell types with naturally rapid turnover (skin, hair and nails, the gastrointestinal tract and the bone marrow) are usually most severely affected. Carriership of germline polymorphisms conferring susceptibility to DNA damage because of lower efficiency of recognition and/or repair may result in higher risk for toxicity after genotoxic treatments.
It is quite difficult to choose between the potential therapeutic options for a particular cancer patient. Even in modern medicine, one could only be truly objective in retrospective when assessing adequacy of a treatment. One could verify that the 'right' therapeutic regimen has been selected of all possible treatments only in the case when both critically important conditions have been fulfilled – the tumour responded to therapy and the adverse effects of the therapy were tolerable. Only the one without the other would be unacceptable, since it may cause needless suffering to human beings that are already very ill.
Knowledge about the individual capacity for DNA repair may be useful in assessment of genotoxic therapies with regard to the eligibility of the particular patient for the particular type of therapy, considering the possible outcomes – expected response to the chosen treatment, expected survival of the patient and the potential adverse effects associated with the therapy. At present, the role of individual repair capacity has only been studied for the most common types of tumours and the most commonly used basic therapeutic regimens, but the field is currently in development.
5.1. Individual repair capacity, eligibility for anticancer therapies and survival in patients with cancer
Publius Vergillis Maro, Aeneid, Book IV.
(She fed within her veins a flame unseen)
Translated by John Dryden (1631–1700).
Eligibility for treatment with genotoxic agents is dependent on the balance between the response to treatment (in terms of destruction or regression of the tumour); the expected survival of the patient (relapse-free as well as overall survival); and the expected toxicity effects (acute as well as delayed). Some treatments that work well in most patients may only have limited effects in some patients, whereas others may have no effect at all. The knowledge whether a patient ought to be started on a certain drug at all may be crucial, as a failed course of treatment may waste valuable time (an important factor in the treatment of all diseases, but especially in advanced cancer) and may significantly worsen the condition of the patient.
Survival in cancer patients (measured in months and years of life after diagnosis) is dependent on many parameters. Some of these are related to characteristics of the tumour – e.g. histological type, localisation; vascularisation of the tumour tissue; differentiation grade of the tumour (poorly, moderately or well differentiated); its immunological and biochemical properties, and others. Others pertain to the organism of the patient – their age; sometimes – their sex; their general condition; the hormonal status (e.g. in estrogen-dependent tumours in women – whether the tumour developed before or after the menopause); the body mass index; some features of the lifestyle and potentially harmful habits (e.g. diet rich in salt, refined sugar and saturated fats, smoking, etc.) and others. Survival (progression-free and overall) may be different for men and women with the same type of cancer. The risks for therapy-related toxicities may also be different between the two sexes. In fact, the risk for developing cancer in the first place may be different between men and women, even when hormone-dependent cancers such as cancer of the mammary gland have been excluded. For example, women have slightly lower lifetime risk for development of colon cancer than men. It is still unclear whether that is because women are more likely to have a 'health-oriented' lifestyle than men (e.g. eating diets lower in fats and richer in fresh fruit and vegetables, smoking less (if at all) and drinking smaller amounts of alcohol) or because of other reasons. Differential attitudes to therapy may also explain (at least partially) why women generally respond better to therapy and have superior survival rates than men – presumably because they are more inclined to stick to a tedious therapeutic schedule than men. Obesity, a universal risk factor for colorectal cancer, was found to be associated with a significant (about 40%) increase of the risk for colon cancer in men, but lower increase of the risk in women (less than 10%) [
One of the critically important predictors for survival in almost all tumours is the grade of local advancement of the tumour (often determined as size of primary tumour and/or degree of involvement of adjacent tissues) and whether involvement of organs and tissues distant from the primary tumour (metastases) are present at the time of diagnosis. Local tissue involvement may be manageable, but usually, the more advanced (either locally or metastatically) a tumour is, the less therapeutic options are available and the poorer the prognosis for the patient.
Individual capacity for repair of DNA damage may influence virtually all factors with roles in the constitution of survival rates: the sensitivity of the tumour to anticancer treatments and the risk of development of resistance mechanisms that may allow the cancer cell to avoid, overcome or compensate for genotoxic damage inflicted by anticancer therapy; and therapy-associated toxicity (tissue- and organ-specific or overall). Usually, the more damage a genotoxic compound causes to the tumour cells, the better the response to the treatment is in terms of tumour shrinkage and obstruction of its blood vessels. One could expect that good response to treatment (expressed as objective tumour regression or, at least, slowing down of cancer progression) would be associated with higher survival rates. This is not, however, always valid, as the effects of the treatment may be significantly compromise the health and the well-being of the patient to the point of being life-threatening. Eventually, the patient may die because of treatment-related adverse effects and not because of progression of the tumour. Usually, DNA polymorphisms decreasing the capacity for repair of DNA damage and maintenance of the genome integrity are associated with better response to treatment in cancer patients. The variant alleles of most polymorphisms in DNA repair genes are often associated with lowering the capacity for recognition and/or repair of DNA damage (although there are many exceptions). Thus, carriership of the variant allele may be associated with increased risk for certain cancers or other diseases and conditions in healthy individuals, whereas the same allele may be associated with better response to therapy and/or superior survival in cancer patients treated with genotoxic agents. Sometimes, however, paradoxical responses may occur, with the polymorphic variant conferring higher capacity for DNA repair but, at the same time, being associated with superior survival in anticancer treatment (see below). What the response to therapy might be and how the survival rates might be shaping, is also dependent on the type of the tumour, the genotoxic agent being used, and other factors pertaining specifically to the patient.
The risk for induction of drug resistance during treatment is also a factor in a patient's survival. The number of treatment options typically becomes more and more limited and the prognosis worsens after the tumour has become resistant to the first-line treatment, as the second-line and third-line anticancer agents may not be that efficient and/or safe to use as these to which the tumour had already become resistant were at first. How long would it be before the cancer cells become capable of overcoming genotoxic damage is dependent on the individual repair capacity, although some types of tumours are more prone to development of resistance to anticancer drugs than others. There are also tumours that are a priori resistant to most modern genotoxic treatments (e.g. inflammatory breast cancer), but these are rare. Most cancer cells that develop resistance to anticancer agents have previously been sensitive or very sensitive to them (for more information, see Chapter XIII – DNA repair and carcinogenesis).
Individual repair capacity and eligibility for different anticancer therapies
The assessment of eligibility for treatment with anticancer drugs is still often based on purely empirical grounds. If the drug in question has known potential for causing severe adverse effects, then patients that are elderly, already frail, or have a pre-existing condition that may be severely exacerbated by treatment with the prospective agent/s may be evaluated as ineligible for the particular treatment on the basis of their physical condition only. Other empirical factors, including lifestyle and habits are also sometimes taken into account. For example, the chemotherapeutic erlotinib, a tyrosine kinase inhibitor suppressing EGFR-related signalling in tumours of epithelial origin (non-small-cell lung cancer and, sometimes, pancreatic and colorectal cancer), was found to be significantly less efficient in smokers, because of accelerated metabolic clearance of the drug [
The biochemical properties of the tumour may also give an indication about whether the patient is eligible for a certain type of treatment. For example, a triple-negative breast tumour(expressing neither the estrogen receptor alpha nor the progesterone receptor or the HER2 receptor) is not likely to respond to estrogen receptor-targeted or HER2-targeted therapies. Therefore, patients with triple-negative tumours (e.g. invasive breast cancer) are usually started on genotoxic therapy without preliminary trials of antiestrogens (e.g. tamoxifen) and herceptin, which would very likely be ineffective [
One of the basic factors in the assessment whether the patient is eligible for treatment with a particular type of anticancer agents is the p53 status of the tumour – specifically, whether the tumour expresses p53 or not, and if it does, whether the p53 is wildtype or a cancer-specific isoform. About 50% of human tumours carry alterations in the TP53 locus, which is usually associated with poorer prognosis for the patient [
Therapies based on p53-activation will only work, however, if the tumour cells are capable of expression of wildtype 53. Patients with tumours that have lost the p53 expression altogether or express a cancer-specific isoform would not be eligible for p53-based treatments, as no significant beneficial effects could be expected. For example, the p53 status is crucially important in assessment of eligibility for different types of treatments in chronic lymphocytic leukemia (CLL). 5–10% of the patients with CLL have a deletion of the 17p genomic region, including the TP53 locus. The patients with intact 17p are eligible for various genotoxic treatments (alkylating agents, e.g. cyclophosphamide; DNA synthesis inhibitors, e.g. fludarabine, and others) that may produce long-lasting remissions. Patients with 17p deletions, however, are likely to benefit more from treatments based on working principles other than induction of the p53-dependent pathways, such as antibodies (alemtuzumab), immunomodulators (lenalidomide), CDK inhibitors (flavopiridol) and steroids [
Determination of levels of the expression of the protein factor ERCC1 may aid the identification of patients eligible for therapy with platinum derivatives. Patients with low or undetectable levels of ERCC1 protein in tumour tissue may exhibit better responses to platinum-based chemotherapy [
In a large study from 2008, among 25 DNA polymorphisms in genes coding for proteins of DNA repair, maintenance of genome integrity and progression through the cell cycle, several were associated with predictably poorer responses to treatment in patients with advanced lung cancer treated with platinum derivatives: the rs1800975 polymorphism in the 5′-UTR of the XPA gene; XPC ins83; XPD Lys751Gln; CCNH Val270Ala; RAD23B Ala249Val; and the ERCC1 C8092A 3'-UTR polymorphism [
Individual repair capacity and survival of patients with cancer
Survival of patients in anticancer therapy may vary greatly, depending on the type of the tumour, the degree of local and/or systemic advancement and factors pertaining to the individual patient. Individual capacity for repair of DNA damage is also a crucial factor. At present, best studied with regard to patient survival after anticancer therapy are polymorphisms in the TP53, XPA, XPC, XPD, XPG and the ERCC1 genes.
As was already mentioned, the presence of wildtype TP53 gene copies in the tumour may be a significant factor determining survival in patients with chronic lymphocytic leukemia. In 20–30% of all patients with CLL it may present as indolent disease, with a prolonged clinical course (up to 10–20 years) and requiring specific treatment only in the late stages or not at all. In patients with 17p deletions, however, the clinical course may be typical of an aggressive tumour resistant to chemotherapy. Deletion of the TP53 locus is associated with more aggressive course and, respectively, with shorter survival, in multiple myeloma [
The role of the common TP53 Pro72Arg polymorphism as a potential survival-modifying factor in various cancers has been extensively studied, but the results have been contradictory at best. It was already mentioned that carriers of Arg72 alleles (specifically, the Arg/Arg homozygotes) may be at selective advantage in some cancers– e.g. some forms of hereditary colorectal cancer [
Carriership of the duplication variant in intron 3 of the TP53 gene may be associated with poorer prognosis in patients with non-small cell lung cancer [
Homozygous carriership of the XPA gene variant rs1800975 in patients with advanced lung cancer treated with platinum-based regimens is associated with shorter survival [
Genotypes containing at least one deletion allele by the XPC ins83 polymorphism(associated with 'normal' repair capacity) may be associated with poorer prognosis in patients with advanced lung cancer treated with platinum derivatives [
The effect of the Lys751Gln polymorphism in the XPD on the survival of patients with non-small cell lung cancer to chemotherapy with platinum derivatives varies in different reports. For example, the Gln allele was associated with poorer survival in the study of Wu et al., [
Carriership of the XPD 312Asn allele in patients with non-small cell lung cancer may confer poorer response to platinum-based regimens and shorter survival than the Asp312 variant [
Carriership of the 399Gln allele in the XRCC1 may also be associated with shorter survival in patients with NSCLC treated with platinum agents [
Heterozygocity by the ERCC1 T19007C (rs11615) polymorphism may be associated with higher 5-year survival in patients treated with combined radio- and chemotherapy for squamous carcinoma of the oesophagus, compared with either of homozygous genotypes [
The His1104Asp variant of the ERCC5 gene was found to be associated with poorer survival in both non-small cell and small cell lung cancer [
The ERCC1 gene has been particularly well studied with regard to patient survival in anticancer therapy. Absence or drastic reduction of ERCC1 expression (at mRNA as well as protein level) was repeatedly reported to be associated with better response to chemotherapy with platinum derivatives and increased overall survival [
The C allele of the synonymous (Asn-Asn) C-to-T substitution at codon 118 of the ERCC1 gene, associated with differential mRNA levels, conferred longer survival in patients with advanced colorectal carcinoma treated with platinum derivatives [
The Lys259Thr polymorphism in the ERCC1 gene was found to be associated with longer overall survival and better response to therapy in patients with refractory multiple myeloma treated with thalidomide [
Carriership of the variant allele of the XRCC1 Arg194Trp polymorphism may indicate better response to treatment with platinum derivatives in patients with advanced non-small cell lung cancer [
In the studies of Li et al. [
5.2. Individual repair capacity and the risk for development of resistance to genotoxic therapies
Cells with near-normal or higher-than-normal capacity for repair of DNA damage may repair therapy-induced genotoxic DNA damage efficiently and may, therefore, be less sensitive (resistant) to genotoxic treatments. When higher-than-normal repair capacity is established by the genetic background of the cell, the tumour may be a priori resistant to treatment. In most cases, however, tumour cells acquire the ability to repair DNA damage caused by anticancer treatments de novo. Strategies for sensitisation of cancer cells by decreasing their capacity to repair the damage inflicted by anticancer treatment and/or the progression in the cell cycle are currently in intensive development. Usually, these strategies are based on inactivation of key protein/s acting in induction of cell cycle arrest and repair of damage in actively dividing cells. Among the common target proteins are, for example, ATM (inactivated by compounds such as KU-55933 and KU59403) [
Platinum-based regimens (where the platinum derivative is used as a single agent or combined with other drugs) are used very often for treatment of solid tumours – mainly because of the high response rates, comparable only to anthracycline-based regimens. Therefore, it is not surprising that patients treated with platinum derivatives are enrolled in clinical studies more often than patients treated with other types of genotoxic drugs. Since platinum complexes are not metabolised or degraded in any other specific fashion (e.g. enzymatic), resistance to platinum-based regimens is dependent on mechanisms such as sequestering the active substance, routing it out of the cell or making it inactive or unavailable before it has found its target. Development of resistance to them is, however, dependent also on the capacity for repair of DNA damage. There may be significant variance in the capacity of tumour cells to repair adducts in DNA caused by treatment with platinum derivatives. High levels of mRNA and protein of the factors of NER ERCC1 and XPD were reported to be associated with resistance to cisplatin therapy in some tumours (e.g. non-small-cell lung cancer) [
Deletion of both somatic TP53 copies in multiple myeloma is a predictor for resistance to genotoxic therapy with 6-mercaptopurine (a pyrimidine analogue) or melphalan (an alkylating agent) [
5.3. Individual repair capacity and the risk of toxicity of anticancer therapies
(Never take the antidote before the poison)
Latin proverb
Individual tolerance to anticancer therapy is a rather complex issue. Generally, the more aggressive a therapy, the better the chances for eradication of the tumour. Treatment with genotoxic agents, however, is always associated with risk of toxicity. Genotoxic treatments are targeted at rapidly cycling cells, but affect, albeit to a lesser degree, all cells capable of division. Cells with a naturally rapid turnover (hair bulbs, skin and mucosa, blood-forming tissue) are especially vulnerable. During or after treatments with genotoxic agents, some patients may become so ill that they may drop out of treatment, or, rarely, may die because of severe inhibition of the normal functioning of healthy cells (e.g. agranulocytopenia, severe skin inflammation, mucositis, severe gastrointestinal disturbances, etc.). Therapy-related adverse effects in tissues that are naturally slow cyclers (cardiotoxicity, neurotoxicity) are usually related to polymorphisms in genes coding for enzymes or other molecules functioning in detoxification of the active compound/s of the drug (cytochrome P450, glutathione S-transferases, UGT1A1, and others). Toxic effects are anticipated in anticancer therapy and the goal is not to prevent their occurrence altogether (which is virtually impossible at present), but to decrease their severity and/or make their effects more tolerable, whenever possible.
The prevalence and the severity of toxic effects related to anticancer therapy may greatly vary depending on the type of the therapeutic agent/s used; but also on the individual characteristics of the patient. The latter include features such as age, general condition, body mass index; lifestyle and habits (e.g. smoking–best studied in lung cancer patients that continue smoking after diagnosis); the genetic background of the patient, including polymorphisms in genes coding for products functioning in DNA repair and maintenance of genome integrity as well as other inherited polymorphisms (for example, in genes coding for factors acting in pro- and anti-inflammatory signalling and tissue renewal).
There is a single common factor that increases the risk for acute toxicity in virtually all types of anticancer treatments, and in children and adults alike – namely, obesity [
Individual repair capacity may play a crucial role in the assessment of the risk for toxicity of anticancer therapies [reviewed in detail in
Carriership of the XRCC1 Arg399Gln and the XRCC3 Thr241Met polymorphisms may predispose to radiation-induced subcutaneous fibrosis and telangiectasias after radiotherapy [
The synonymous polymorphism Asp568Asp in the LIG4 gene, Asp711Asp in the XPD gene, the 5′-untranslated region polymorphism in XRCC3 gene and the Val219Ile polymorphism in the MLH1 gene were found to be associated with late rectal and/or bladder toxicity in patients treated with radiotherapy for prostate cancer [
Significant association with severe (grade 3–4) toxicity (myelosuppression/dysphagia) was identified for 12 out of 21 studied polymorphisms in genes coding for products functioning in DNA repair, maintenance of genome integrity and the control of the progression in the cell cycle [
Carriership of the XRCC1 399Gln and APE1 148Glu alleles in breast cancer patients treated with radiotherapy was associated with reduced risk for moist desquamation of irradiated normal skin[
The TP53 Pro72Arg polymorphism may be associated with risk for development of atypical vascular lesions at sites of radiotherapy for breast-conserving therapy for breast carcinoma [
Heterozygous carriership of mutations associated with ataxia-telangiectasia when inherited in homozygous state, or 'neutral' polymorphisms in the ATM gene may be associated with high toxicity in patients treated with ionising radiation [
Some of the polymorphic gene variants that are associated with superior survival after anticancer treatments (usually, these are variants conferring lower capacity for DNA repair) are also factors in the risk for toxicity due to genotoxic treatments. This could only be expected, as the healthy cells in the patient have the same baseline repair capacity and would, therefore, also suffer more damage in genotoxic treatments. For example, XPD haplotype by the markers Asp312Asn and Lys751Gln may be associated with risk for high-grade neutropenia. In patients with advanced NSCLC treated with platinum derivatives, carriership of the XPD Asp312/Lys751 haplotype (associated with better response to therapy and significantly increased one-year and overall survival) was also associated with greater risk for very severe (grade 4) neutropenia compared with carriers of XPD Asn312/Gln751 haplotype [
The relationship between improved survival rates and risk for therapy-associated toxicity may, however, be more complicated. Alleles that confer higher repair capacity may sometimes be associated with severe toxic effects due to anticancer therapy. For example, among patients with non-small cell lung cancer treated with platinum derivatives, increased risk for high-grade (grade 3–4) gastrointestinal toxicity was not seen in carriers of the C allele of the ERCC1 gene (associated with lower ERCC1 transcript stability and lower protein levels), but in carriers of the A allele [
Toxicity effects may sometimes even be used as phenotypic markers for response to genotoxic treatments. For example, the already mentioned EGFR inhibitor erlotinib causes papulopustular eruption (rash) in some patients. In about 10% of all patients, the rash is severe (grade >2). Studies show that the occurrence and the grade of the rash may correlate with the degree of response to therapy and patient survival in advanced cancer, with one-year survival being between 2 and 3 times higher in patients that have had rashes grade 2–4 during treatment with erlotinib than in patients with low-grade rash or no rash at all [
Of course, one cannot rely solely on the assessment of adverse effects when it comes to evaluation of response. Nevertheless, phenotypic markers of this type may be a rapid and relatively reliable way to judge whether a drug trial is going well or not.
6. Commonly used methods for assessment of capacity for DNA repair
Communion with her visible forms, she speaks
a various language.
William Cullen Bryant, Thanatopsis (1817)
Research related to DNA repair may be fundamental, targeted at elucidation of the basic principles governing the detection and repair of damage and maintenance of genome integrity, the different repair pathways and mechanisms; or applied, involving definition and validation of biomolecular markers for assessment of individual repair capacity and their use in research and clinical settings, individualisation of therapies, etc. Both types of research need reliable methods for measurement of efficiency of repair of DNA damage. There are a legion of possible logical variants for planning and implementation of experiments in the field of assessment of DNA repair. The basic methodologies may involve assessment of repair of specific types of lesions; methodologies that are independent of the type of lesion; approaches that evaluate the rates of repair in specific model systems; methodologies that work in vitro only or in vivo (in situ), in specific genomic regions or globally throughout the genome.
It is difficult to measure directly the rates of DNA repair, as the types of damage may be very different from one another, even when caused in controlled conditions (in these cases the dose of the damaging agent is typically calibrated), and the capacity of the DNA repair machinery for removal of different types of lesions may show significant intra-individual variation. Therefore, the capacity for repair of DNA damage is usually measured by indirect methods. Generally, a specific type of damage is introduced in the DNA of living cells (or in a DNA fragment or construct that is subsequently introduced into living cells), then the cells are allowed to repair the damage. The time intervals during which the cells carry out the repair may vary and the process of repair may be allowed to proceed normally or may be impeded in some way (e.g. under conditions of deficiency of a specific nucleotide or enzyme activity). After the time for repair is due, the number of unrepaired lesions or their proportion to the overall amount of damage immediately after the phase of introduction of damage are elicited (by various methods), recorded and compared to the levels of unrepaired damage in control (untreated) cells, or the number or the proportion of unrepaired lesions when the type of damage is different, or the same type of damage is introduced in other genomic regions, or the same type of damage is repaired by other mechanisms. The occurrence of repair is usually demonstrated by the recovery of the initial DNA as it was before the damage occurred (its primary sequence and/or its structure), or by restoration of its functionality (e.g. may serve as a template for DNA polymerase). Some of the methodologies are applicable only to one type of lesion (e.g. base mismatches), others are more versatile with regard to the type of lesion and its location in the genome.
At the moment, research on individual capacity for cell and tissue renewal is still a very young field that is, however, rapidly expanding. Biomarkers in this field are not numerous yet, and markers for cell proliferation are not always applicable, as stimulation of cell cycle in some cells may not result in cell division, but, rather, may trigger apoptosis. At the moment, telomerase activity, telomere length and rate of attrition of telomere ends are the most commonly used phenotypic markers for the capacity for cell and tissue renewal.
Some of the specific methods that are currently used to measure rates of DNA repair, individual repair capacity and capacity for cell and tissue renewal are outlined below.
6.1. Assessment of the capacity for repair of DNA damage
Mutagen challenge assay
The term usually pertains to double-strand breaks, but essentially all assays that include controlled treatments with DNA damaging agents and monitoring of the appearance and/or the disappearance of the lesions (such as the Ames test, T4 endonuclease V test, the assay for unscheduled synthesis of DNA. etc.) are, by definition, mutagen challenge assays. The classic assay is based on measuring the amount of induced chromatid breaks at metaphase in cultured cells (most often – human peripheral blood lymphocytes) after exposure to various mutagens during the S–G2 phase of the cell cycle [
Alkaline elution
Alkaline elution was initially developed for assessment of capacity for repair of single-strand breaks in DNA [
Briefly, the cells are treated with the damaging agent, and then allowed to repair the damage for fixed interval/s of time. Labelled nucleotide/s is/are supplied to be incorporated in the newly synthesised DNA. After the time allowed for repair is due, the cells are harvested and their DNA is extracted by any of the various methods for isolation of high-molecular weight DNA, in order to yield a population of long DNA fragments containing the unrepaired lesions. Alternatively, if the studied DNA fragments are cloned in vectors propagated in prokaryotic cells, cells growing in liquid medium are harvested by centrifugation and DNA is extracted by alkaline lysis or any other of the methods for extraction and purification of plasmid DNA. If the cells grow on solid medium in Petri dishes, the colonies are blotted onto nylon filters, lysed and the DNA is fixed onto the filters. If the type of damage is different from single-strand breaks or abasic sites in DNA, at this step the extracted DNA is treated in order to convert the lesion to an abasic site or a single-strand break. For example, if the damaging agents cause modification of nucleotides, the extracted DNA is treated with specific glycosylases that recognise the sites of damage and convert them to abasic sites. The latter may subsequently be converted to single-strand breaks (e.g. by hydrolysis at alkaline pH). The DNA carrying the single-strand breaks is then denatured and immobilised onto solid supports. Short DNA fragments fix poorly onto the support and may be eluted in alkaline buffers. Therefore, the more unrepaired lesions there were in DNA, the greater the number of single-strand breaks and, respectively, the proportion of eluted short DNA fragments. Provided that the amount of starting DNA is known, the levels of the DNA retained onto the solid support and in the eluate may be calculated by measuring and quantitating the levels of the label. This reflects the amount of breaks in DNA and, respectively, the amounts of unrepaired damage. Control samples are run in parallel to the test samples, with all samples being treated in identical way. The repair capacity of the tested samples is calculated as the ratio of the amount of DNA breaks in the test samples to the amount of breaks in the control cells.
Т4 endonuclease V method
The T4 endonuclease V method [
Briefly, the test cells are irradiated with UV and allowed to repair the lesions for fixed interval/s of time. Control unirradiated cell are run in parallel. The cells are then harvested, the DNA is extracted (preferably, by methods yielding high-molecular weight DNA) and subjected to hydrolysis with one or more restriction endonucleases so as to produce fragments with suitable length. The preparation of hydrolysed DNA is treated with T4 endonuclease V, and then electrophoresed in alkaline buffer (in order to denature the DNA). The separated fragments are blotted and fixed onto nylon membrane. Southern hybridisation with a labelled DNA probe specific to the studied region/s is then carried out and the resulting image is digitised and analysed (e.g. by densitometry) in order to assess the relative amount of signal generated by the fragments hybridising with the labelled probe in the test cells and in the control cells. At sites where the thymine dimers in DNA had not been repaired, the treatment with T4endoV generates single-strand breaks, eventually resulting in a population of shorter DNA fragments. The latter migrate at faster rates during electrophoresis when run in parallel with DNA, in which the damage had been repaired, producing distinct electrophoretic profiles. After blotting, very short fragments (presumably resulting from inefficient repair of DNA) are poorly retained by the membrane, and the hybridisation signal from cells that have sustained unrepaired damage is decreased when compared to undamaged cells or cells that have had the damage repaired. Thus, lower repair capacity is associated with weaker signal after visualisation, shifted towards fragments with lower molecular weight, whereas cells with higher repair capacity generate signal with higher intensity, shifted towards the fragments with higher molecular weight.
The T4endoV method is very sensitive (not surprising, as it is based on enzyme-substrate recognition), but is applicable to one type of lesion only. It is semiquantitative, as the hybridisation signal (e.g. from an autoradiographic image) may be digitised and the ratio of the levels of signal in test cells compared to control cells may be calculated. For short DNA regions, only one labelled probe might suffice, but when the studied region is longer, several different probes must be used.
Pulsed field electrophoresis
Pulsed field electrophoresis is usually used for assessment of repair of double-strand breaks (e.g. resulting from ionising radiation and/or radiomimetic chemicals) [
F = e-LB,
where F is the ratio between the amount of probe hybridised to DNA from a test sample and the amount of probe hybridised to the control cells, L is the length of the fragments to which the probe hybridises (in [Kb]), and B is the number of double-strand breaks per restriction fragment.
Denaturing gel electrophoresis
Capacity for repair of crosslinking damage (specifically, interstrand links) may be assessed using denaturing gel electrophoresis [
P0 = e-X,
where Р0 is the relative amount (percentage) of DNA that exhibits electrophoretic behaviour characteristic of single-strand DNA (no crosslinking) and X is the average number of crosslinks per restriction fragment [
Measurement of unscheduled (non-replicative) synthesis of DNA
Regardless of the type of damage and the exact mechanism, DNA repair is always associated with synthesis of DNA. Therefore, measuring the rate of DNA synthesis may be used to assess the rate of DNA repair. Many methods for labelling and detection of newly synthesised nucleic acids have been developed, allowing quantitative assessment of the rate of incorporation of labelled nucleotides, and, respectively, the rate of DNA synthesis. The bulk of DNA synthesis in a cell, however, is related to replication, and the synthesis of DNA associated with repair activities constitutes only a very small part of the overall DNA synthesis. In actively dividing cells it might be virtually impossible to separate the relative contribution of the one type of synthesis and the other. Of course, the test cells may be sorted according to phase of cell cycle to ensure that no cells that are actively replicating their DNA are included in the sample. Sometimes, however, the sample size is small and/or the cells that are replicatively quiescent are very rare within the sample. A possible workaround is to block the replicative DNA synthesis but not repair-associated synthesis, or at least to make sure that repair-related synthesis is not as severely suppressed as the replicative synthesis. The most commonly used agent for blocking replicative DNA synthesis is hydroxyurea [
There are variations to the methodology, but basically it includes the exposition of test and control cells to the damaging agent followed immediately by treatment with hydroxyurea in order to suppress replicative DNA synthesis. Then the cells are allowed to repair the damage and the newly synthesised DNA (presumably resulting exclusively from the repair activities) is labelled by culturing in nutrient medium containing labelled nucleotide/s. The cells are harvested, and then their DNA is extracted, purified and cleaved into fragments of suitable length. The population of fragments is hybridised against a set of probes and the amount of label-containing DNA hybridised against the probes is determined by different methods (depending on the label) – e.g. by immunochemistry [
PCR-based methods for assessment of capacity for repair of DNA damage
Since the polymerase chain reaction was introduced into routine laboratory practice with the work of Saiki et al. [
Another PCR-based methodology for analysis of capacity for repair of DNA damage is based on the fact that virtually all types of DNA damage present a challenge to DNA polymerases (except in the specific cases of translesion replication). Damaged DNA templates cannot usually be copied by the DNA polymerases routinely used in in vitro amplification. Briefly, after the test cells have been treated with genotoxic agents and allowed to repair the damage, their DNA is extracted and set for in vitro amplification using specific primers. Control cells (untreated) are usually run in parallel. The primers and/or the nucleosidetriphosphates in the reaction may be labelled in order to follow the rate of synthesis, or a fluorescent double-strand specific dye (e.g. SYBR green) may be added to the reaction mix. The amount of the synthesised PCR product is quantitated (usually in real-time). The rate of repair is measured as the ratio of the yield of full-length double-strand PCR product in test cells and the yield of product in control cells [
Assessment of repair capacity by transformation assays
This methodology is based on the routine methods for transformation of prokaryotic cells with plasmid constructs. The cells are treated with genotoxic agents (most commonly, causing double-strand breaks) and allowed time to repair the damage. Then the DNA is extracted, subjected to hydrolysis with restriction endonucleases and fragments of desired length or specific DNA sequences are collected and purified by any of the routine methods. The fragments are then ligated into a linearised vector and prokaryotic (usually, E. coli) cells are transformed. Presumably, only perfect circular molecules are capable of transforming prokaryotic cells. If the damage has been repaired correctly, the cells would be transformed and the reporter gene (usually, a marker for resistance to an antibiotic) would be expressed in the transformants, allowing them to grow on a selective medium [
Immunochemical methods for assessment of capacity for DNA repair
Immunochemical methods may be used for direct observation of the occurrence of the damage event and its repair (e.g. appearance and subsequent disappearance of a modified base) or for indirect monitoring of the process of repair via the molecules and the supramolecular complexes that recognise and repair the damage. The former may be pretty straightforward –creating antibodies specific to the damaged molecular species, labelling the antibodies with a readily detectable label, and monitoring the occurrence and the distribution of the signal. For example, monoclonal antibodies have been created against virtually all types of modified bases and photoproducts in DNA [
Mass spectrometry
Mass spectrometry is by far the most specific direct method for assessment of capacity for repair of DNA damage. It allows sensitive and specific detection of the presence of virtually all types of DNA lesions. Basically, the cells are exposed to the damaging agents and allowed variable intervals of time for repair as usual. Control cells (treated but not allowed time for repair and untreated cells) are run in parallel. DNA is then extracted, fragmented, analysed and the results are compared to the results obtained with the control cells in order to monitor the kinetics of appearance and disappearance of the lesions. Mass spectrometry has been successfully applied in studies of repair of oxidised bases, DNA dimers, DNA adducts, etc. [
Assessment of DNA repair capacity using cell-free systems
The methodology is based on introduction of known type of lesions into DNA fragments and allowing the damage to be repaired in cell-free systems (lysates, extracts). Different types of DNA lesions and different damage burden per DNA molecule may be analysed. Assessment of repair efficiency itself is carried out by additional methods (e.g. measuring non-replicative DNA synthesis by the rate of inclusion of labelled nucleotide precursors).
Analysis of repair in cell-free systems has the advantage that it may be used in assessment of repair of base mismatches. This particular type of damage poses difficulties to virtually all other conventional methods for assessment of DNA repair. It is not a DNA modification per se, as both mismatched nucleotides are normal compounds of DNA and cannot be recognised by glycosylases or damage-specific antibodies. As the DNA fragments carrying damage are usually cloned into vectors, presence of unrepaired mismatches may be detected by sequencing the plasmid insert before and after incubation in cell-free systems for fixed intervals of time [
Host cell reactivation (HCR)
HCR methodology is based on analysis of repair of a reporter sequence in living eukaryotic cells. Briefly, damage is introduced in a specific reporter gene of a vector (various types, virus as well as plasmid). The latter is then used for transfection of eukaryotic cells and the cells are allowed to repair the damage for fixed intervals of time. In the presence of unrepaired damage in the reporter gene its capacity for being transcribed and expressed would presumably be impaired; therefore, the rate of recovery of the expression of the reporter gene is monitored and compared to the rate of control cells. The reporter (marker) sequences used in HCR may vary – chloramphenicol acetyltransferase, luciferase, GFP and other fluorescent proteins, and possibly others [
HCR is applicable for virtually all types of damage, possibly with the exception of double-strand breaks, as they would cause linearisation of the circular DNA of plasmid vectors and circularised virus vectors. However, as assessment of efficiency of repair of double-strand breaks may be implemented by other methods (e.g. pulsed-field electrophoresis, immunochemical methods, etc.), this is not an issue. Moreover, some years ago, a variation of HCR was developed that allowed assessment by HCR of repair of double-strand breaks by homologous recombination and NHEJ, [
HCR was reported to had been successfully used for assessment of individual capacity for DNA repair in clinically healthy people and patients with cancer [
Comet assay (single-cell electrophoresis) for assessment of strand breaks and apoptosis
Comet assay is another method based on electrophoresis, but the electric force is applied not on molecules, but on whole cells. Comet assay analyses the differential patterns of electrophoretic profiles of single cells after genotoxic treatments producing single-strand and double-strand breaks and other types of damage that may be converted to DNA breaks. For example, one of the variants of the comet assay utilises specific repair endonucleases to convert DNA lesions to single-strand breaks (e.g. endonuclease III (nth) may be used for assessment of presence of oxidised pyrimidine bases) [reviewed in
The cells are treated with the damaging agent, and then allowed time for repair. Control (untreated) cells are usually run in parallel. The cells are immobilised in a porous medium (e.g. low-gelling point agarose), then subjected to lysis and denaturation of DNA. Electrophoresis is then carried out onto glass slides and the DNA is stained by an intercalating double-strand specific dye (ethidium bromide, SYBR green, etc.). The resulting image may be observed under fluorescent microscope. The DNA of untreated control cells (presumably intact or containing very small baseline amount of damage) is normally visualised as a fluorescent halo. If the DNA of the test cells contains unrepaired breaks, it becomes fragmented and migrates in a specific fashion, generating an image resembling a comet's tail, with the amount of DNA in the tail corresponding to the amount of damage (that, is, the number of breaks) that the cells had sustained [
A variation of the comet assay methodology exists, allowing assessment of repair of crosslinking damage. The working principle is basically the same, with slight variations, but the end image is interpreted in a different manner. After the cells had been treated with crosslinking agents and allowed to repair, they are subjected to treatments introducing strand breaks. Control cells (not treated with crosslinking agents, but subjected to treatment introducing strand breaks) are also run in parallel. From this point onward, the protocol is essentially the same as in 'classic' comet assay. In cells that have sustained crosslinks and have not managed to repair them during the allotted time, after the electrophoresis, fragments containing damage sites would be impeded in their migration into the 'comet's tail'. In control cells, there would presumably be no unrepaired crosslinks to keep the DNA fragments bound together; therefore, DNA would migrate easily into the comet's tail. The relative amount of DNA damage is then inversely proportional to the amount of DNA in the 'comet's tail' and is calculated as the ratio of the amount of DNA in the comet's tail in test cells and in control cells [
Cell-free extracts may also be used in comet assay. Briefly, the test extract is incubated with damaged substrate DNA, and the incisions due to repair activity are detected as DNA breaks. In this case, the amount of DNA in the tail corresponds to the incision activity of the extract. Adequate controls are always run in parallel and the capacity for repair of damage calculated as the ratio of the amount of DNA in the comet's tail in test cells and in control cells.
The comet assay has many advantages and is a commonly used laboratory technique. It is applicable to various cell types and is sufficiently sensitive to detect even low levels of DNA damage. It is also rapid, cost-effective, requires small numbers of cells per analysis and does not usually call for specialised personnel training [reviewed in
6.2. Assessment of the capacity for self-renewal in cell populations
Capacity for self-renewal of cell populations is an important parameter in the studies of ageing, cell differentiation, assessment of properties of different types of tumour cells (proliferative capacity, potential for genomic instability, etc.) and many other areas of biomedical science, fundamental as well as applied. Telomere length, telomere attrition rate and/or telomerase activity are useful markers for the capacity for self-renewal of cell populations. In unsynchronised cell populations (as almost all native eukaryotic cell populations are), the mean telomere length is comprised of the relative contributions of the telomere length of the individual cells. Therefore, the end result would be strongly dependent on the heterogeneity of the cell population and its reproducibility would be dependent on the dynamics of this heterogeneity. Sometimes even small differences may have diagnostic or prognostic meaning, but only in specific context and/or in specific periods. For example, in patients with history of myocardial infarction, the difference between telomere lengths of granulocytes and lymphocytes in the same blood sample has been shown to reflect the degree of impairment of haematopoietic progenitor cells [
The major methods used for measuring telomere length in cell populations are the mean terminal restriction fragment assay (MRF, often referred to by the descriptive name of the routine technique it is based upon – Southern hybridisation), quantitative PCR and flow cytometry/flow FISH. The major method for assessment of telomerase activity is currently TRAP (Telomeric Repeat Amplification Protocol). There are other methods as well, but they are not commonly used, such as Single Telomere Length Analysis (STELA). The working principles of major methods and their advantages and disadvantages are outlined briefly below [reviewed in detail in
Mean terminal restriction fragment method (MRF)
Historically first among the methods to determine telomere length, the MRF methodology is still the gold standard in the assays for determination of telomere length [
Quantitative PCR for measuring telomere length
Quantitative PCR (qPCR) is based on monitoring the increase in the amount of double-stranded amplification product during a PCR reaction, measured by increase of fluorescence using a standard curve. There are, however, numerous variations of the technique. Among the most popular methodologies for qPCR-based measurement of telomere length is the T/S method (Telomere/Single Copy Gene ratio) [
Quantitative PCR is rapid, requires relatively small amounts of PCR-grade DNA and may easily be implemented in high-throughput format. It is, however, sensitive to changes in the amount of template and, as in all PCR-based assays; there is an inherent risk of cross-contamination.
Flow cytometry/Flow FISH
In its barebones version the method is based on hybridisation of telomeric DNA of fixed and permeabilised cells with a fluorescently labelled probe – usually, a peptide-nucleotide probe (PNA) with sequence complementary to the repeated unit in telomere DNA [
This method for determination of telomere length is recently gaining in popularity, as it is relatively rapid and makes use of samples containing a small number of cells. Major limiting factors are the requirement for designated equipment and specifically trained personnel and the availability of PNA probes.
Single Telomere Length Analysis (STELA)
This is a somewhat exotic assay based on ligation of a linker complementary to the telomeric repeats to the DNA extracted from the test samples, followed by in vitro amplification using primers specific for the linker and chromosome-specific primers for the subtelomeric regions, and subsequent Southern hybridisation with probes representing the sequence from the subtelomeric regions [
In order to monitor the rate of telomere attrition, which is a more reliable marker for the capacity for cell and tissue renewal than mean telomere length alone, telomere length is usually measured at fixed intervals of time. In tissues with preserved telomerase activity, the latter may be measured instead of telomere attrition rate, or the two methods may be used simultaneously to obtain higher reliability of the results.
TRAP analysis
The TRAP (Telomeric Repeat Amplification Protocol) assay [
Acknowledgеments
This research was supported by Grant No. DFNI-B01/2 at the National Science Fund, Ministry of Education and Science of Republic of Bulgaria.
References
- Pero RW, Bryngelsson C, Mitelman F, Kornfält R, Thulin T, Norden A. Interindividual variation in the responses of cultured human lymphocytes to exposure from DNA damaging chemical agents: interindividual variation to carcinogen exposure. Mutat Res. 1978;53(3):327–41.
- Pero RW, Ostlund C. Direct comparison, in humans resting lymphocytes, of the inter-individual variations in unscheduled DNA synthesis induced by N-acetoxy-2-acetylaminofluorene and ultraviolet irradiation. Mutat Res. 1980;73(2):349–61.
- Dayer P, Balant L, Fabre J. The genetic control of drug oxidation in the liver. Int J Clin Pharmacol Res. 1983;3(6):421–5.
- Guengerich FP, Beaune PH, Umbenhauer DR, Churchill PF, Bork RW, Dannan GA, et al. Cytochrome P-450 enzymes involved in genetic polymorphism of drug oxidation in humans. Biochem Soc Trans. 1987;15(4):576–8.
- Shimada T, Watanabe J, Kawajiri K, Sutter TR, Guengerich FP, Gillam EM, et al. Catalytic properties of polymorphic human cytochrome P450 1B1 variants. Carcinogenesis. 1999;20(8):1607–13.
- Calabrese EJ. Sex differences in susceptibility to toxic industrial chemicals. Br J Ind Med. 1986;43(9):577–9.
- Petkova R, Chakarov S, Ganev V. Genetic Bases For Predisposition To Common Multifactorial Disease In Man. Part II. Biotechnol Biotechnol Eq. 2007;21(4):385–92.
- Petkova R, Chakarov S, Ganev V. Genetic Bases For Predisposition To Common Multifactorial Disease In Man. Part I. Biotechnol Biotechnol Eq. 2007;21(3):286–93.
- Hanova M, Stetina R, Vodickova L, Vaclavikova R, Hlavac P, Smerhovsky Z, et al. Modulation of DNA repair capacity and mRNA expression levels of XRCC1, hOGG1 and XPC genes in styrene-exposed workers. Toxicol Appl Pharmacol. 2010;248(3):194–200.
- Budworth H, Snijders AM, Marchetti F, Mannion B, Bhatnagar S, Kwoh E, et al. DNA repair and cell cycle biomarkers of radiation exposure and inflammation stress in human blood. Borgmann K, editor. PLoS One. Public Library of Science; 2012;7(11):e48619.
- Chicheva Z, Chelenkova P, Petkova R, Chakarov S. Children Of The Sun, Children Of The Moon – A Mini-Panel For Assessment Of Inter-Individual Variation Between The Capacity Of Healthy Individuals To Repair Everyday Genotoxic Insults. Biotechnol Biotechnol Eq. 2012;26(4):3142–7.
- Forrester HB, Li J, Hovan D, Ivashkevich AN, Sprung CN. DNA repair genes: alternative transcription and gene expression at the exon level in response to the DNA damaging agent, ionizing radiation. PLoS One. 2012;7(12):e53358.
- Chelenkova P, Petkova R, D' Ascanio I, Zhelev N, Chakarov St. In sickness and in health: a set of markers for individual repair capacity in risk assessment, monitoring and prognosis of human disease. Curr Opin Biotechnol. July 2013; 24(Supplement 1):S105.
- Chelenkova P, Petkova R, Chamova T, Zheliazkova-Glaveeva S, Tournev I, Chakarov S. Homozygous Carriership Of The Wildtype Allele Of The XPCins83 Polymorphism May Be An Independent Protective Factor Against Cerebrovascular Incidents In The Bulgarian Population. Comptes rendus l'academie Bulg des Sci. 2014;67(2):263–8.
- Plappert UG, Stocker B, Fender H, Fliedner TM. Changes in the repair capacity of blood cells as a biomarker for chronic low-dose exposure to ionizing radiation. Environ Mol Mutagen. 1997;30(2):153–60.
- Vodicka P, Kumar R, Stetina R, Sanyal S, Soucek P, Haufroid V, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25(5):757–63.
- Fracasso ME, Doria D, Franceschetti P, Perbellini L, Romeo L. DNA damage and repair capacity by comet assay in lymphocytes of white-collar active smokers and passive smokers (non- and ex-smokers) at workplace. Toxicol Lett. 2006;167(2):131–41.
- Hemminki K. Re: Markers of DNA Repair and Susceptibility to Cancer in Humans: an Epidemiologic Review. J Natl Cancer Inst. 2000;92(18):1536–7.
- Mohrenweiser HW, Wilson DM, Jones IM. Challenges and complexities in estimating both the functional impact and the disease risk associated with the extensive genetic variation in human DNA repair genes. Mutat Res. 2003;526(1-2):93–125.
- Huppke P, Maier EM, Warnke A, Brendel C, Laccone F, Gärtner J. Very mild cases of Rett syndrome with skewed X inactivation. J Med Genet. 2006;43(10):814–6.
- Horibata K, Iwamoto Y, Kuraoka I, Jaspers NGJ, Kurimasa A, Oshimura M, et al. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci U S A. 2004;101(43):15410–5.
- Skjelbred CF, Saebø M, Wallin H, Nexø BA, Hagen PC, Lothe IMB, et al. Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma, in a Norwegian cohort: a case control study. BMC Cancer. 2006;6:67.
- Abdel-Rahman SZ, Soliman AS, Bondy ML, Omar S, El-Badawy SA, Khaled HM, et al. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159(1):79–86.
- Oscarson M, Ingelman-Sundberg M. CYPalleles: a web page for nomenclature of human cytochrome P450 alleles. Drug Metab Pharmacokinet. 2002;17(6):491–5.
- Zhou S-F, Liu J-P, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41(2):89–295.
- Reszka E, Czekaj P, Adamska J, Wasowicz W. Relevance of glutathione S-transferase M1 and cytochrome P450 1A1 genetic polymorphisms to the development of head and neck cancers. Clin Chem Lab Med. 2008;46(8):1090–6.
- Sergentanis TN, Economopoulos KP. Four polymorphisms in cytochrome P450 1A1 (CYP1A1) gene and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;122(2):459–69.
- Marichalar-Mendia X, Rodriguez-Tojo MJ, Acha-Sagredo A, Rey-Barja N, Aguirre-Urizar JM. Oral cancer and polymorphism of ethanol metabolising genes. Oral Oncol. 2010;46(1):9–13.
- Tao L, Xiang Y-B, Chan KK, Wang R, Gao Y-T, Yu MC, et al. Cytochrome P4501A2 phenotype and bladder cancer risk: The Shanghai bladder cancer study. Int J Cancer. 2012;130(5):1174–83.
- Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25(11):1679–91.
- Leskelä S, Jara C, Leandro-García LJ, Martínez A, García-Donas J, Hernando S, et al. Polymorphisms in cytochromes P450 2C8 and 3A5 are associated with paclitaxel neurotoxicity. Pharmacogenomics J. 2011;11(2):121–9.
- Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22(47):7369–75.
- Bolt H, Thier R. Relevance of the Deletion Polymorphisms of the Glutathione S-Transferases GSTT1 and GSTM1 in Pharmacology and Toxicology. Curr Drug Metab. Bentham Science Publ Ltd; 2006;7(6):613–28.
- Clapper ML. Genetic polymorphism and cancer risk. Curr Oncol Rep. 2000;2(3):251–6.
- Ye Z, Song H. Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. Eur J Cancer. 2005;41(7):980–9.
- Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–600.
- Kudo T, Asano J, Shimizu T, Nanashima N, Fan Y, Akita M, et al. Different susceptibility to peroxisome proliferator-induced hepatocarcinogenesis in rats with polymorphic glutathione transferase genes. Cancer Sci. 2006;97(8):703–9.
- Shimizu T, Fan Y, Yamana D, Miura T, Nanashima N, Yamada T, et al. Glutathione S-transferase A4 is a positive marker for rat hepatic foci induced by clofibrate and genotoxic carcinogens. Cancer Sci. 2010;101(5):1093–8.
- Allan JM, Wild CP, Rollinson S, Willett E V, Moorman A V, Dovey GJ, et al. Polymorphism in glutathione S-transferase P1 is associated with susceptibility to chemotherapy-induced leukemia. Proc Natl Acad Sci U S A. 2001;98(20):11592–7.
- Russo A, Francelin PR, Galbiatti ALS, Raposo LS, Maníglia JV, Pavarino EC, et al. Association between GSTP1, GSTM1 and GSTT1 polymorphisms involved in xenobiotic metabolism and head and neck cancer development. Mol Biol Rep. 2013;40(7):4181–8.
- Zusterzeel P. Glutathione S-transferase isoenzymes in decidua and placenta of preeclamptic pregnancies. Obstet Gynecol. 1999;94(6):1033–8.
- Zusterzeel PL, Visser W, Peters WH, Merkus HW, Nelen WL, Steegers EA. Polymorphism in the glutathione S-transferase P1 gene and risk for preeclampsia. Obstet Gynecol. 2000;96(1):50–4.
- Zusterzeel PLM. Paternal contribution to the risk for pre-eclampsia. J Med Genet. 2002;39(1):44–5.
- Cibeira MT, de Larrea CF, Navarro A, Díaz T, Fuster D, Tovar N, et al. Impact on response and survival of DNA repair single nucleotide polymorphisms in relapsed or refractory multiple myeloma patients treated with thalidomide. Leuk Res. 2011;35(9):1178–83.
- De Mattia E, Toffoli G. C677T and A1298C MTHFR polymorphisms, a challenge for antifolate and fluoropyrimidine-based therapy personalisation. Eur J Cancer. 2009;45(8):1333–51.
- Wilcken B, Bamforth F, Li Z, Zhu H, Ritvanen A, Renlund M, et al. Geographical and ethnic variation of the 677C>T allele of 5,10 methylenetetrahydrofolate reductase (MTHFR): findings from over 7000 newborns from 16 areas world wide. J Med Genet. 2003;40(8):619–25.
- Cohen V, Panet-Raymond V, Sabbaghian N, Morin I, Batist G, Rozen R. Methylenetetrahydrofolate Reductase Polymorphism in Advanced Colorectal Cancer: A Novel Genomic Predictor of Clinical Response to Fluoropyrimidine-based Chemotherapy. Clin Cancer Res. 2003;9(5):1611–5.
- Ji M, Xu B, Jiang J-T, Wu J, Li X-D, Zhao W-Q, et al. Relationship between glutathione S-transferase P1 (GSTP1), X-ray repair cross complementing group 1 (XRCC1) and 5,10-methylenetetrahydrofolate reductase (5,10-MTHFR) gene polymorphisms and response to chemotherapy in advanced gastric cancer. Onkologie. 2013;36(6):335–40.
- Blank S, Rachakonda S, Keller G, Weichert W, Lordick F, Langer R, et al. A retrospective comparative exploratory study on two Methylentetrahydrofolate Reductase (MTHFR) polymorphisms in esophagogastric cancer: the A1298C MTHFR polymorphism is an independent prognostic factor only in neoadjuvantly treated gastric cancer patient. BMC Cancer. 2014;14:58.
- Tiseo M, Giovannetti E, Tibaldi C, Camerini A, Di Costanzo F, Barbieri F, et al. Pharmacogenetic study of patients with advanced non-small cell lung cancer (NSCLC) treated with second-line pemetrexed or pemetrexed-carboplatin. Lung Cancer. 2012;78(1):92–9.
- Toffoli G, Russo A, Innocenti F, Corona G, Tumolo S, Sartor F, et al. Effect of methylenetetrahydrofolate reductase 677C-->T polymorphism on toxicity and homocysteine plasma level after chronic methotrexate treatment of ovarian cancer patients. Int J Cancer. 2003;103(3):294–9.
- Eissa DS, Ahmed TM. C677T and A1298C polymorphisms of the methylenetetrahydrofolate reductase gene: effect on methotrexate-related toxicity in adult acute lymphoblastic leukaemia. Blood Coagul Fibrinolysis. 2013;24(2):181–8.
- Tanaka Y, Manabe A, Nakadate H, Kondoh K, Nakamura K, Koh K, et al. Methylenetetrahydrofolate reductase gene haplotypes affect toxicity during maintenance therapy for childhood acute lymphoblastic leukemia in Japanese patients. Leuk Lymphoma. 2013;
- Hagleitner MM, Coenen MJH, Aplenc R, Patiño-Garcia A, Chiusolo P, Gemmati D, et al. The role of the MTHFR 677C>T polymorphism in methotrexate-induced liver toxicity: a meta-analysis in patients with cancer. Pharmacogenomics J. 2014;14(2):115–9.
- Loganayagam A, Arenas Hernandez M, Corrigan A, Fairbanks L, Lewis CM, Harper P, et al. Pharmacogenetic variants in the DPYD, TYMS, CDA and MTHFR genes are clinically significant predictors of fluoropyrimidine toxicity. Br J Cancer. 2013;108(12):2505–15.
- Bastaki M, Huen K, Manzanillo P, Chande N, Chen C, Balmes JR, et al. Genotype-activity relationship for Mn-superoxide dismutase, glutathione peroxidase 1 and catalase in humans. Pharmacogenet Genomics. 2006;16(4):279–86.
- Ruiz-Sanz JI, Aurrekoetxea I, Matorras R, Ruiz-Larrea MB. Ala16Val SOD2 polymorphism is associated with higher pregnancy rates in in vitro fertilization cycles. Fertil Steril. 2011;95(5):1601–5.
- Paludo FJ de O, Picanço JB, Fallavena PRV, Fraga L da R, Graebin P, Nóbrega O de T, et al. Higher frequency of septic shock in septic patients with the 47C allele (rs4880) of the SOD2 gene. Gene. 2013;517(1):106–11.
- Sandström J, Nilsson P, Karlsson K, Marklund SL. 10-fold increase in human plasma extracellular superoxide dismutase content caused by a mutation in heparin-binding domain. J Biol Chem. 1994;269(29):19163–6.
- Juul K, Tybjaerg-Hansen A, Marklund S, Heegaard NHH, Steffensen R, Sillesen H, et al. Genetically reduced antioxidative protection and increased ischemic heart disease risk: The Copenhagen City Heart Study. Circulation. 2004;109(1):59–65.
- Kinnula VL, Crapo JD. Superoxide dismutases in the lung and human lung diseases. Am J Respir Crit Care Med. 2003;167(12):1600–19.
- Liu G, Zhou W, Park S, Wang LI, Miller DP, Wain JC, et al. The SOD2 Val/Val genotype enhances the risk of nonsmall cell lung carcinoma by p53 and XRCC1 polymorphisms. Cancer. 2004;101(12):2802–8.
- Young RP, Hopkins R, Black PN, Eddy C, Wu L, Gamble GD, et al. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax. 2006;61(5):394–9.
- Tamai M, Furuta H, Kawashima H, Doi A, Hamanishi T, Shimomura H, et al. Extracellular superoxide dismutase gene polymorphism is associated with insulin resistance and the susceptibility to type 2 diabetes. Diabetes Res Clin Pract. 2006;71(2):140–5.
- Möllsten A, Jorsal A, Lajer M, Vionnet N, Tarnow L. The V16A polymorphism in SOD2 is associated with increased risk of diabetic nephropathy and cardiovascular disease in type 1 diabetes. Diabetologia. 2009;52(12):2590–3.
- Tian C, Fang S, Du X, Jia C. Association of the C47T polymorphism in SOD2 with diabetes mellitus and diabetic microvascular complications: a meta-analysis. Diabetologia. 2011;54(4):803–11.
- Rosta K, Molvarec A, Enzsöly A, Nagy B, Rónai Z, Fekete A, et al. Association of extracellular superoxide dismutase (SOD3) Ala40Thr gene polymorphism with pre-eclampsia complicated by severe fetal growth restriction. Eur J Obstet Gynecol Reprod Biol. 2009;142(2):134–8.
- Andreassen CN, Alsner J, Overgaard M, Overgaard J. Prediction of normal tissue radiosensitivity from polymorphisms in candidate genes. Radiother Oncol. 2003;69(2):127–35.
- Ahn J, Ambrosone CB, Kanetsky PA, Tian C, Lehman TA, Kropp S, et al. Polymorphisms in genes related to oxidative stress (CAT, MnSOD, MPO, and eNOS) and acute toxicities from radiation therapy following lumpectomy for breast cancer. Clin Cancer Res. 2006;12(23):7063–70.
- Kamatani T, Yamamoto T, Yoneda K, Osaki T. Polymorphic mutations of the Mn-SOD gene in intact human lymphocytes and oral squamous cell carcinoma cell lines. Biochem Cell Biol. 2003;81(1):43–50.
- Monari M, Trinchero A, Calabrese C, Cattani O, Serrazanetti GP, Foschi J, et al. Superoxide dismutase in gastric adenocarcinoma: is it a clinical biomarker in the development of cancer? Biomarkers. 11(6):574–84.
- Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32(5):651–62.
- Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, Krynetskaia NF, et al. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58(4):694–702.
- Candelaria M, Taja-Chayeb L, Arce-Salinas C, Vidal-Millan S, Serrano-Olvera A, Dueñas-Gonzalez A. Genetic determinants of cancer drug efficacy and toxicity: practical considerations and perspectives. Anticancer Drugs. 2005;16(9):923–33.
- Coulthard S, Hogarth L. The thiopurines: an update. Invest New Drugs. 2005;23(6):523–32.
- Fabre MA, Jones DC, Bunce M, Morris PJ, Friend PJ, Welsh KI, et al. The impact of thiopurine S-methyltransferase polymorphisms on azathioprine dose 1 year after renal transplantation. Transpl Int. 2004;17(9):531–9.
- Kurzawski M, Dziewanowski K, Gawrońska-Szklarz B, Domański L, Droździk M. The impact of thiopurine s-methyltransferase polymorphism on azathioprine-induced myelotoxicity in renal transplant recipients. Ther Drug Monit. 2005;27(4):435–41.
- Diasio RB, Johnson MR. Dihydropyrimidine dehydrogenase: its role in 5-fluorouracil clinical toxicity and tumor resistance. Clin Cancer Res. 1999;5(10):2672–3.
- Harris BE, Carpenter JT, Diasio RB. Severe 5-fluorouracil toxicity secondary to dihydropyrimidine dehydrogenase deficiency. A potentially more common pharmacogenetic syndrome. Cancer. 1991;68(3):499–501.
- Van Kuilenburg AB, Vreken P, Abeling NG, Bakker HD, Meinsma R, Van Lenthe H, et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum Genet. 1999;104(1):1–9.
- Ezzeldin H, Diasio R. Dihydropyrimidine dehydrogenase deficiency, a pharmacogenetic syndrome associated with potentially life-threatening toxicity following 5-fluorouracil administration. Clin Colorectal Cancer. 2004;4(3):181–9.
- Maring JG, Groen HJM, Wachters FM, Uges DRA, de Vries EGE. Genetic factors influencing pyrimidine-antagonist chemotherapy. Pharmacogenomics J. 2005;5(4):226–43.
- Baskin Y, Amirfallah A, Unal OU, Calibasi G, Oztop I. Dihydropyrimidine Dehydrogenase 85T>C Mutation Is Associated With Ocular Toxicity of 5-Fluorouracil: A Case Report. Am J Ther. 2013;25 July 20.
- Burchell B, Nebert DW, Nelson DR, Bock KW, Iyanagi T, Jansen PL, et al. The UDP glucuronosyltransferase gene superfamily: suggested nomenclature based on evolutionary divergence. DNA Cell Biol. 1991;10(7):487–94.
- Toffoli G, Cecchin E. Uridine diphosphoglucuronosyl transferase and methylenetetrahydrofolate reductase polymorphisms as genomic predictors of toxicity and response to irinotecan-, antifolate- and fluoropyrimidine-based chemotherapy. J Chemother. 2004;16 Suppl 4:31–5.
- Marsh S, McLeod HL. Pharmacogenetics of irinotecan toxicity. Pharmacogenomics. Future Medicine Ltd London, UK; 2004;5(7):835–43.
- Cheng L, Li M, Hu J, Ren W, Xie L, Sun Z-P, et al. UGT1A1*6 polymorphisms are correlated with irinotecan-induced toxicity: a system review and meta-analysis in Asians. Cancer Chemother Pharmacol. 2014;73(3):551–60.
- Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006;54(3):267–83.
- De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631(2):101–10.
- López-Cima MF, González-Arriaga P, García-Castro L, Pascual T, Marrón MG, Puente XS, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162.
- Karahalil B, Bohr VA, Wilson DM. Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk. Hum Exp Toxicol. 2012;31(10):981–1005.
- Mellon I, Hock T, Reid R, Porter PC, States JC. Polymorphisms in the human xeroderma pigmentosum group A gene and their impact on cell survival and nucleotide excision repair. DNA Repair (Amst). 2002;1(7):531–46.
- Wu X, Zhao H, Wei Q, Amos CI, Zhang K, Guo Z, et al. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis. 2003;24(3):505–9.
- Khan SG, Metter EJ, Tarone RE, Bohr VA, Grossman L, Hedayati M, et al. A new xeroderma pigmentosum group C poly(AT) insertion/deletion polymorphism. Carcinogenesis. 2000;21(10):1821–5.
- Marín MS, López-Cima MF, García-Castro L, Pascual T, Marrón MG, Tardón A. Poly (AT) polymorphism in intron 11 of the XPC DNA repair gene enhances the risk of lung cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(11 Pt 1):1788–93.
- Blankenburg S, König IR, Moessner R, Laspe P, Thoms K-M, Krueger U, et al. Assessment of 3 xeroderma pigmentosum group C gene polymorphisms and risk of cutaneous melanoma: a case-control study. Carcinogenesis. 2005;26(6):1085–90.
- Casson AG, Zheng Z, Evans SC, Veugelers PJ, Porter GA, Guernsey DL. Polymorphisms in DNA repair genes in the molecular pathogenesis of esophageal (Barrett) adenocarcinoma. Carcinogenesis. 2005;26(9):1536–41.
- Kietthubthew S, Sriplung H, Au WW, Ishida T. Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health. 2006;209(1):21–9.
- Yang P-W, Hsieh C-Y, Kuo F-T, Huang P-M, Hsu H-H, Kuo S-W, et al. The survival impact of XPA and XPC genetic polymorphisms on patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2013;20(2):562–71.
- Lee GY, Jang J-S, Lee SY, Jeon H-S, Kim KM, Choi JE, et al. XPC polymorphisms and lung cancer risk. Int J Cancer. 2005;115(5):807–13.
- Dai Q-S, Hua R-X, Zhang R, Huang Y-S, Hua Z-M, Yun CT, et al. Poly (AT) deletion/insertion polymorphism of the XPC gene contributes to urinary system cancer susceptibility: a meta-analysis. Gene. 2013;528(2):335–42.
- Qiu L, Wang Z, Shi X, Wang Z. Associations between XPC polymorphisms and risk of cancers: A meta-analysis. Eur J Cancer. 2008;44(15):2241–53.
- Zhu Y, Yang H, Chen Q, Lin J, Grossman HB, Dinney CP, et al. Modulation of DNA damage/DNA repair capacity by XPC polymorphisms. DNA Repair (Amst). 2008;7(2):141–8.
- Vodicka P, Kumar R, Stetina R, Musak L, Soucek P, Haufroid V, et al. Markers of individual susceptibility and DNA repair rate in workers exposed to xenobiotics in a tire plant. Environ Mol Mutagen. 2004;44(4):283–92.
- Laczmanska I, Gil J, Karpinski P, Stembalska A, Trusewicz A, Pesz K, et al. Polymorphism in nucleotide excision repair gene XPC correlates with bleomycin-induced chromosomal aberrations. Environ Mol Mutagen. 2007;48(8):666–71.
- Tricot O, Mallat Z, Heymes C, Belmin J, Lesèche G, Tedgui A. Relation between endothelial cell apoptosis and blood flow direction in human atherosclerotic plaques. Circulation. 2000;101(21):2450–3.
- Pérez-Mayoral J, Pacheco-Torres AL, Morales L, Acosta-Rodríguez H, Matta JL, Dutil J. Genetic polymorphisms in RAD23B and XPC modulate DNA repair capacity and breast cancer risk in Puerto Rican women. Mol Carcinog. 2013;52 Suppl 1:E127–38.
- Tomoda T, Nouso K, Sakai A, Ouchida M, Kobayashi S, Miyahara K, et al. Genetic risk of hepatocellular carcinoma in patients with hepatitis C virus: a case control study. J Gastroenterol Hepatol. 2012;27(4):797–804.
- Sturgis EM, Dahlstrom KR, Spitz MR, Wei Q. DNA repair gene ERCC1 and ERCC2/XPD polymorphisms and risk of squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2002;128(9):1084–8.
- Jiang J, Zhang X, Yang H, Wang W. Polymorphisms of DNA repair genes: ADPRT, XRCC1, and XPD and cancer risk in genetic epidemiology. Methods Mol Biol. 2009;471:305–33.
- Yin Z, Su M, Li X, Li M, Ma R, He Q, et al. ERCC2, ERCC1 polymorphisms and haplotypes, cooking oil fume and lung adenocarcinoma risk in Chinese non-smoking females. J Exp Clin Cancer Res. 2009;28:153.
- Mandal RK, Gangwar R, Mandhani A, Mittal RD. DNA repair gene X-ray repair cross-complementing group 1 and xeroderma pigmentosum group D polymorphisms and risk of prostate cancer: a study from North India. DNA Cell Biol. 2010;29(4):183–90.
- Chang C-H, Wang R-F, Tsai R-Y, Wu H-C, Wang C-H, Tsai C-W, et al. Significant association of XPD codon 312 single nucleotide polymorphism with bladder cancer susceptibility in Taiwan. Anticancer Res. 2009;29(10):3903–7.
- Gangwar R, Ahirwar D, Mandhani A, Mittal RD. Influence of XPD and APE1 DNA repair gene polymorphism on bladder cancer susceptibility in north India. Urology. 2009;73(3):675–80.
- Unal M, Güven M, Batar B, Ozaydin A, Sarici A, Devranoğlu K. Polymorphisms of DNA repair genes XPD and XRCC1 and risk of cataract development. Exp Eye Res. 2007;85(3):328–34.
- Padma G, Mamata M, Reddy KRK, Padma T. Polymorphisms in two DNA repair genes (XPD and XRCC1)--association with age related cataracts. Mol Vis. 2011;17:127–33.
- Woelfelschneider A, Popanda O, Lilla C, Linseisen J, Mayer C, Celebi O, et al. A distinct ERCC1 haplotype is associated with mRNA expression levels in prostate cancer patients. Carcinogenesis. 2008;29(9):1758–64.
- Chen P, Wiencke J, Aldape K, Kesler-Diaz A, Miike R, Kelsey K, et al. Association of an ERCC1 Polymorphism with Adult-Onset Glioma. Cancer Epidemiol Biomarkers Prev. 2000;9(8):843–7.
- Zhang L, Wang J, Xu L, Zhou J, Guan X, Jiang F, et al. Nucleotide excision repair gene ERCC1 polymorphisms contribute to cancer susceptibility: a meta-analysis. Mutagenesis. 2012;27(1):67–76.
- Cui Y, Morgenstern H, Greenland S, Tashkin DP, Mao J, Cao W, et al. Polymorphism of Xeroderma Pigmentosum group G and the risk of lung cancer and squamous cell carcinomas of the oropharynx, larynx and esophagus. Int J Cancer. 2006;118(3):714–20.
- Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27(3):560–7.
- Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103(6):1864–9.
- Salmanoglu M, Kucukardali Y, Kucukodaci Z, Fenercioglu A, Solmazgul E, Onem Y, et al. Prevalence of the DNA repair enzyme-NEIL1 gene mutation in patients with type 2 diabetes in the Turkish population. J Endocrinol Invest. 2012;35(4):401–6.
- Park M-H, Kwak SH, Kim KJ, Go MJ, Lee H-J, Kim K-S, et al. Identification of a genetic locus on chromosome 4q34-35 for type 2 diabetes with overweight. Exp Mol Med. 2013;45:e7.
- Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S, et al. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet. 2007;121(2):233–42.
- Figueroa JD, Malats N, Rothman N, Real FX, Silverman D, Kogevinas M, et al. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28(8:1788–93.
- Zhang H, Xu Y, Zhang Z, Li L. The hOGG1 Ser326Cys polymorphism and prostate cancer risk: a meta-analysis of 2584 cases and 3234 controls. BMC Cancer. 2011;11:391.
- Zhang Y, Zhang L, Song Z, Sun DL, Liu HR, Fu S Bin, et al. Genetic polymorphisms in DNA repair genes OGG1, APE1, XRCC1, and XPD and the risk of age-related cataract. Ophthalmology. 2012;119(5):900–6.
- Audebert M, Chevillard S, Levalois C, Gyapay G, Vieillefond A, Klijanienko J, et al. Alterations of the DNA repair gene OGG1 in human clear cell carcinomas of the kidney. Cancer Res. 2000;60(17):4740–4.
- Li Q, Huang L, Rong L, Xue Y, Lu Q, Rui Y, et al. hOGG1 Ser326Cys polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Cancer Sci. 2011;102(6):1123–7.
- Tsai C-C, Wu S-B, Cheng C-Y, Kao S-C, Kau H-C, Chiou S-H, et al. Increased oxidative DNA damage, lipid peroxidation, and reactive oxygen species in cultured orbital fibroblasts from patients with Graves' ophthalmopathy: evidence that oxidative stress has a role in this disorder. Eye (Lond). 2010;24(9):1520–5.
- Tanrikulu S, Doğru-Abbasoğlu S, Ozderya A, Ademoğlu E, Karadağ B, Erbil Y, et al. The 8-oxoguanine DNA N-glycosylase 1 (hOGG1) Ser326Cys variant affects the susceptibility to Graves' disease. Cell Biochem Funct. 2011;29(3):244–8.
- Savas S, Kim DY, Ahmad MF, Shariff M, Ozcelik H. Identifying functional genetic variants in DNA repair pathway using protein conservation analysis. Cancer Epidemiol Biomarkers Prev. 2004;13(5):801–7.
- Takanami T, Nakamura J, Kubota Y, Horiuchi S. The Arg280His polymorphism in X-ray repair cross-complementing gene 1 impairs DNA repair ability. Mutat Res. 2005;582(1-2):135–45.
- Hsieh Y-Y, Chang C-C, Chen S-Y, Chen C-P, Lin W-H, Tsai F-J. XRCC1 399 Arg-related genotype and allele, but not XRCC1 His107Arg, XRCC1 Trp194Arg, KCNQ2, AT1R, and hOGG1 polymorphisms, are associated with higher susceptibility of endometriosis. Gynecol Endocrinol. 2012;28(4):305–9.
- Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM. Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28(20):3871–9.
- Jing B, Wang J, Chang W-L, Li B, Chen J, Niu Y-J. Association of the polymorphism of APE1 gene with the risk of prostate cancer in Chinese Han population. Clin Lab. 2013;59(1-2):163–8.
- Kang H, Dai Z, Ma X, Ma L, Jin Y, Liu X, et al. A genetic variant in the promoter of APE1 gene (-656 T>G) is associated with breast cancer risk and progression in a Chinese population. Gene. 2013;531(1):97–100.
- Wang M, Chu H, Wang S, Wang M, Wang W, Han S, et al. Genetic variant in APE1 gene promoter contributes to cervical cancer risk. Am J Obstet Gynecol. 2013;209(4):360.e1–7.
- Thyagarajan B, Lindgren B, Basu S, Nagaraj S, Gross MD, Weisdorf DJ, et al. Association between genetic variants in the base excision repair pathway and outcomes after hematopoietic cell transplantations. Biol Blood Marrow Transplant. 2010;16(8):1084–9.
- Arora M, Lindgren B, Basu S, Nagaraj S, Gross M, Weisdorf D, et al. Polymorphisms in the base excision repair pathway and graft-versus-host disease. Leukemia. 2010;24(8):1470–5.
- Wang M, Chu H, Zhang Z, Wei Q. Molecular epidemiology of DNA repair gene polymorphisms and head and neck cancer. J Biomed Res. 2013;27(3):179–92.
- Santos RA, Teixeira AC, Mayorano MB, Carrara HHA, Andrade JM, Takahashi CS. DNA repair genes XRCC1 and XRCC3 polymorphisms and their relationship with the level of micronuclei in breast cancer patients. Genet Mol Biol. 2010;33(4):637–40.
- Varon R, Reis A, Henze G, Einsiedel HG v., Sperling K, Seeger K. Mutations in the Nijmegen Breakage Syndrome Gene (NBS1) in Childhood Acute Lymphoblastic Leukemia (ALL). Cancer Res. 2001;61(9):3570–2.
- Shimada H, Shimizu K, Mimaki S, Sakiyama T, Mori T, Shimasaki N, et al. First case of aplastic anemia in a Japanese child with a homozygous missense mutation in the NBS1 gene (I171V) associated with genomic instability. Hum Genet. 2004;115(5):372–6.
- Desjardins S, Beauparlant JC, Labrie Y, Ouellette G, Durocher F. Variations in the NBN/NBS1 gene and the risk of breast cancer in non-BRCA1/2 French Canadian families with high risk of breast cancer. BMC Cancer. 2009;9:181.
- Roddam PL, Rollinson S, O'Driscoll M, Jeggo PA, Jack A, Morgan GJ. Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination. J Med Genet. 2002;39(12):900–5.
- Girard P-M, Kysela B, Härer CJ, Doherty AJ, Jeggo PA. Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms. Hum Mol Genet. 2004;13(20):2369–76.
- Long X-D, Zhao D, Wang C, Huang X-Y, Yao J-G, Ma Y, et al. Genetic polymorphisms in DNA repair genes XRCC4 and XRCC5 and aflatoxin B1-related hepatocellular carcinoma. Epidemiology. 2013;24(5):671–81.
- Lin Z-H, Chen J-C, Wang Y-S, Huang T-J, Wang J, Long X-D. DNA repair gene XRCC4 codon 247 polymorphism modified diffusely infiltrating astrocytoma risk and prognosis. Int J Mol Sci. 2014;15(1):250–60.
- Campbell PT, Curtin K, Ulrich CM, Samowitz WS, Bigler J, Velicer CM, et al. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58(5):661–7.
- Yu J-H, Bigler J, Whitton J, Potter JD, Ulrich CM. Mismatch repair polymorphisms and colorectal polyps: hMLH1-93G>A variant modifies risk associated with smoking. Am J Gastroenterol. 2006;101(6):1313–9.
- Park SH, Lee GY, Jeon H-S, Lee SJ, Kim KM, Jang SS, et al. -93G-->A polymorphism of hMLH1 and risk of primary lung cancer. Int J Cancer. 2004;112(4):678–82.
- Lee K-M, Choi J-Y, Kang C, Kang CP, Park SK, Cho H, et al. Genetic polymorphisms of selected DNA repair genes, estrogen and progesterone receptor status, and breast cancer risk. Clin Cancer Res. 2005;11(12):4620–6.
- Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20(4-5):269–76.
- Medeiros F, Lindor NM, Couch FJ, Highsmith WE. The germline MLH1 K618A variant and susceptibility to Lynch syndrome-associated tumors. J Mol Diagn. 14(3):264–73.
- Perera S, Bapat B. The MLH1 variants p.Arg265Cys and p.Lys618Ala affect protein stability while p.Leu749Gln affects heterodimer formation. Hum Mutat. 2008;29(2):332.
- Conde J, Silva SN, Azevedo AP, Teixeira V, Pina JE, Rueff J, et al. Association of common variants in mismatch repair genes and breast cancer susceptibility: a multigene study. BMC Cancer. 2009;9:344.
- Jin G, Wang H, Hu Z, Liu H, Sun W, Ma H, et al. Potentially functional polymorphisms of EXO1 and risk of lung cancer in a Chinese population: A case-control analysis. Lung Cancer. 2008;60(3):340–6.
- Bau D-T, Wang H-C, Liu C-S, Chang C-L, Chiang S-Y, Wang R-F, et al. Single-nucleotide polymorphism of the Exo1 gene: association with gastric cancer susceptibility and interaction with smoking in Taiwan. Chin J Physiol. 2009;52(6):411–8.
- Hsu N-Y, Wang H-C, Wang C-H, Chiu C-F, Tseng H-C, Liang S-Y, et al. Lung cancer susceptibility and genetic polymorphisms of Exo1 gene in Taiwan. Anticancer Res. 2009;29(2):725–30.
- Wang H-C, Chiu C-F, Tsai R-Y, Kuo Y-S, Chen H-S, Wang R-F, et al. Association of genetic polymorphisms of EXO1 gene with risk of breast cancer in Taiwan. Anticancer Res. 2009;29(10):3897–901.
- Song F, Qureshi AA, Zhang J, Zhan J, Amos CI, Lee JE, et al. Exonuclease 1 (EXO1) gene variation and melanoma risk. DNA Repair (Amst). 2012;11(3):304–9.
- Haghighi MM, Taleghani MY, Mohebbi SR, Vahedi M, Fatemi SR, Zali N, et al. Impact of EXO1 polymorphism in susceptibility to colorectal cancer. Genet Test Mol Biomarkers. 2010;14(5):649–52.
- Gao W-M, Romkes M, Day RD, Siegfried JM, Luketich JD, Mady HH, et al. Association of the DNA repair gene XPD Asp312Asn polymorphism with p53 gene mutations in tobacco-related non-small cell lung cancer. Carcinogenesis. 2003;24(10):1671–6.
- Hou S-M, Ryk C, Kannio A, Angelini S, Fält S, Nyberg F, et al. Influence of common XPD and XRCC1 variant alleles on p53 mutations in lung tumors. Environ Mol Mutagen. 2003;41(1):37–42.
- Gao W-M, Romkes M, Siegfried JM, Luketich JD, Keohavong P. Polymorphisms in DNA repair genes XPD and XRCC1 and p53 mutations in lung carcinomas of never-smokers. Mol Carcinog. 2006;45(11):828–32.
- Smith TR, Liu-Mares W, Van Emburgh BO, Levine EA, Allen GO, Hill JW, et al. Genetic polymorphisms of multiple DNA repair pathways impact age at diagnosis and TP53 mutations in breast cancer. Carcinogenesis. 2011;32(9):1354–60.
- Whibley C, Pharoah PDP, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9(2):95–107.
- Ara S, Lee PS, Hansen MF, Saya H. Codon 72 polymorphism of the TP53 gene. Nucleic Acids Res. 1990;18(16):4961.
- Gemignani F, Moreno V, Landi S, Moullan N, Chabrier A, Gutiérrez-Enríquez S, et al. A TP53 polymorphism is associated with increased risk of colorectal cancer and with reduced levels of TP53 mRNA. Oncogene. 2004;23(10):1954–6.
- Felley-Bosco E, Weston A, Cawley HM, Bennett WP, Harris CC. Functional studies of a germ-line polymorphism at codon 47 within the p53 gene. Am J Hum Genet. 1993;53(3):752–9.
- Dumont P, Leu JI-J, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–65.
- Bulavin D V, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, et al. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999;18(23):6845–54.
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102(6):849–62.
- Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem. 2005;280(25):24245–51.
- Pinto GR, Yoshioka FKN, Silva RLL, Clara CA, Santos MJ, Almeida JRW, et al. Prognostic value of TP53 Pro47Ser and Arg72Pro single nucleotide polymorphisms and the susceptibility to gliomas in individuals from Southeast Brazil. Genet Mol Res. 2008;7(1):207–16.
- Sameer AS, Shah ZA, Syeed N, Banday MZ, Bashir SM, Bhat BA, et al. TP53 Pro47Ser and Arg72Pro polymorphisms and colorectal cancer predisposition in an ethnic Kashmiri population. Genet Mol Res. 2010;9(2):651–60.
- Wu X. p53 Genotypes and Haplotypes Associated With Lung Cancer Susceptibility and Ethnicity. CancerSpectrum Knowl Environ. 2002;94(9):681–90.
- Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two Polymorphic Variants of Wild-Type p53 Differ Biochemically and Biologically. Mol Cell Biol. 1999;19(2):1092–100.
- Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108(2):196–9.
- Frank AK, Leu JI-J, Zhou Y, Devarajan K, Nedelko T, Klein-Szanto A, et al. The codon 72 polymorphism of p53 regulates interaction with NF-{kappa}B and transactivation of genes involved in immunity and inflammation. Mol Cell Biol. 2011;31(6):1201–13.
- Jeong B-S, Hu W, Belyi V, Rabadan R, Levine AJ. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. FASEB J. 2010;24(5):1347–53.
- Beckman G, Birgander R, Själander A, Saha N, Holmberg PA, Kivelä A, et al. Is p53 polymorphism maintained by natural selection? Hum Hered. 44(5):266–70.
- Sergentanis TN, Economopoulos KP. Latitude may modify the effect of TP53 codon 72 polymorphism on cancer risk. Cancer. 2010;116(14):3523.
- Moan J, Grigalavicius M, Baturaite Z, Juzeniene A, Dahlback A. North-South gradients of melanomas and non-melanomas: A role of vitamin D? Dermatoendocrinol. 2013;5(1):186–91.
- Han J, Cox DG, Colditz GA, Hunter DJ. The p53 codon 72 polymorphism, sunburns, and risk of skin cancer in US Caucasian women. Mol Carcinog. 2006;45(9):694–700.
- Krüger S, Bier A, Engel C, Mangold E, Pagenstecher C, von Knebel Doeberitz M, et al. The p53 codon 72 variation is associated with the age of onset of hereditary non-polyposis colorectal cancer (HNPCC). J Med Genet. 2005;42(10):769–73.
- Farrington SM, McKinley AJ, Carothers AD, Cunningham C, Bubb VJ, Sharp L, et al. Evidence for an age-related influence of microsatellite instability on colorectal cancer survival. Int J Cancer. 2002;98(6):844–50.
- Oh J, Kim JW, Lee BE, Jang MJ, Chong SY, Park PW, et al. Polymorphisms of the pri-miR-34b/c promoter and TP53 codon 72 are associated with risk of colorectal cancer. Oncol Rep. 2014;31(2):995–1002.
- Hünten S, Siemens H, Kaller M, Hermeking H. The p53/microRNA network in cancer: experimental and bioinformatics approaches. Adv Exp Med Biol. 2013;774:77–101.
- Son MS, Jang MJ, Jeon YJ, Kim WH, Kwon C-I, Ko KH, et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene. 2013;524(2):156–60.
- Zhou X, Gu Y, Zhang S-L. Association between p53 codon 72 polymorphism and cervical cancer risk among Asians: a HuGE review and meta-analysis. Asian Pac J Cancer Prev. 2012;13(10):4909–14.
- Cherdyntseva N V, Gervas PA, Litvyakov N V, Stakcheeva MN, Ponomaryeva AA, Dobrodeev AY, et al. Age-related function of tumor suppressor gene TP53:contribution to cancer risk and progression. Exp Oncol. 2010;32(3):205–8.
- Zehbe I, Voglino G, Wilander E, Genta F, Tommasino M. Codon 72 polymorphism of p53 and its association with cervical cancer. Lancet. 1999;354(9174):218–9.
- Zehbe I, Voglino G, Wilander E, Delius H, Marongiu A, Edler L, Klimek F, Andersson S, Tommasino M. p53 codon 72 polymorphism and various human papillomavirus 16 E6 genotypes are risk factors for cervical cancer development. Cancer Res. 2001 Jan 15;61(2):608-11.
- Burroni E, Bisanzi S, Sani C, Puliti D, Carozzi F. Codon 72 polymorphism of p53 and HPV type 16 E6 variants as risk factors for patients with squamous epithelial lesion of the uterine cervix. J Med Virol. 2013;85(1):83–90.
- Petkova R, Chelenkova P, Georgieva E, Chakarov S. What's Your Poison? Impact Of Individual Repair Capacity On The Outcomes Of Genotoxic Therapies In Cancer. Part I – Role Of Individual Repair Capacity In The Constitution Of Risk For Late-Onset Multifactorial Disease. Biotechnol Biotechnol Eq. 2013;27(6):4208–16.
- Mitra S, Banerjee S, Misra C, Singh RK, Roy A, Sengupta A, et al. Interplay between human papilloma virus infection and p53 gene alterations in head and neck squamous cell carcinoma of an Indian patient population. J Clin Pathol. 2007;60(9):1040–7.
- Nelson HH, Wilkojmen M, Marsit CJ, Kelsey KT. TP53 mutation, allelism and survival in non-small cell lung cancer. Carcinogenesis. 2005;26(10):1770–3.
- Baynes JW. Role of oxidative stress in development of complications in diabetes. Diabetes. 1991;40(4):405–12.
- Burgdorf KS, Grarup N, Justesen JM, Harder MN, Witte DR, Jørgensen T, et al. Studies of the association of Arg72Pro of tumor suppressor protein p53 with type 2 diabetes in a combined analysis of 55,521 Europeans. Weedon M, editor. PLoS One. Public Library of Science; 2011;6(1):e15813.
- Qu L, He B, Pan Y, Xu Y, Zhu C, Tang Z, et al. Association between polymorphisms in RAPGEF1, TP53, NRF1 and type 2 diabetes in Chinese Han population. Diabetes Res Clin Pract. 2011;91(2):171–6.
- Bonfigli AR, Sirolla C, Testa R, Cucchi M, Spazzafumo L, Salvioli S, et al. The p53 codon 72 (Arg72Pro) polymorphism is associated with the degree of insulin resistance in type 2 diabetic subjects: a cross-sectional study. Acta Diabetol. 2013;50(3):429–36.
- Spitsina E V, Iakunina NI, Chudakova DA, Nikitin AG, Svetlova GN, Soluianova TN, et al. [Association of polymorphous markers Pro72Arg and C(-594)CC OF TP53 gene with diabetic polyneuropathy in patients with type 1 diabetes mellitus living in Moscow]. Mol Biol (Mosk). 41(6):989–93.
- Beiras-Fernandez A, Angele MK, Koutang C, Lohse P, Reichart B, Eifert S. Genetic polymorphisms of TP53 and FAS promoter modulate the progression of coronary artery disease after coronary artery bypass grafting: a gender-specific view. Inflamm Res. 2011;60(5):439–45.
- Gomez-Sanchez JC, Delgado-Esteban M, Rodriguez-Hernandez I, Sobrino T, Perez de la Ossa N, Reverte S, et al. The human Tp53 Arg72Pro polymorphism explains different functional prognosis in stroke. J Exp Med. 2011;208(3):429–37.
- Bonafé M, Salvioli S, Barbi C, Trapassi C, Tocco F, Storci G, et al. The different apoptotic potential of the p53 codon 72 alleles increases with age and modulates in vivo ischaemia-induced cell death. Cell Death Differ. 2004;11(9):962–73.
- Kang H-J, Feng Z, Sun Y, Atwal G, Murphy ME, Rebbeck TR, et al. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A. 2009;106(24):9761–6.
- Feng Z, Zhang C, Kang H-J, Sun Y, Wang H, Naqvi A, et al. Regulation of female reproduction by p53 and its family members. FASEB J. 2011;25(7):2245–55.
- Paskulin D d'Avila, Paixão-Côrtes VR, Hainaut P, Bortolini MC, Ashton-Prolla P. The TP53 fertility network. Genet Mol Biol. 2012;35(4 (suppl)):939–46.
- Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450(7170):721–4.
- Ghafari F, Pelengaris S, Walters E, Hartshorne GM. Influence of p53 and genetic background on prenatal oogenesis and oocyte attrition in mice. Hum Reprod. 2009;24(6):1460–72.
- Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest. 2010;120(3):803–15.
- Corbo RM, Gambina G, Scacchi R. How contemporary human reproductive behaviors influence the role of fertility-related genes: the example of the p53 gene. Singh SR, editor. PLoS One. Public Library of Science; 2012;7(4):e35431.
- Hamilton BE, Ph D, Martin JA, Ventura SJ, Statistics V. National Vital Statistics Reports Births: Preliminary Data for 2010. 2011;60(2):1–26.
- Su Y, Swift M. Mortality rates among carriers of ataxia-telangiectasia mutant alleles. Ann Intern Med. 2000;133(10):770–8.
- Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97(11):813–22.
- Gilad S, Bar-Shira A, Harnik R, Shkedy D, Ziv Y, Khosravi R, et al. Ataxia-telangiectasia: founder effect among north African Jews. Hum Mol Genet. 1996;5(12):2033–7.
- Swift M, Reitnauer PJ, Morrell D, Chase CL. Breast and other cancers in families with ataxia-telangiectasia. N Engl J Med. 1987;316(21):1289–94.
- Morrell D, Chase CL, Swift M. Cancers in 44 families with ataxia-telangiectasia. Cancer Genet Cytogenet. 1990;50(1):119–23.
- Prokopcova J, Kleibl Z, Banwell CM, Pohlreich P. The role of ATM in breast cancer development. Breast Cancer Res Treat. 2007;104(2):121–8.
- Lo Y-L, Hsiao C-F, Jou Y-S, Chang G-C, Tsai Y-H, Su W-C, et al. ATM polymorphisms and risk of lung cancer among never smokers. Lung Cancer. 2010;69(2):148–54.
- Gao Y, Hayes RB, Huang W-Y, Caporaso NE, Burdette L, Yeager M, et al. DNA repair gene polymorphisms and tobacco smoking in the risk for colorectal adenomas. Carcinogenesis. 2011;32(6):882–7.
- Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2(1):41–6.
- Khalil H, Tummala H, Chakarov S, Zhelev N, Lane D. Targeting ATM pathway for therapeutic intervention in cancer. Biodiscovery. Dundee Science Press; 2012;Volume 1.
- Ribeiro HL, Oliveira RTG de, Maia ARS, Sousa JC de, Heredia FF, Magalhães SMM, et al. ATM polymorphism is associated with low risk myelodysplastic syndrome. DNA Repair (Amst). 2013;12(2):87–9.
- Bay JO, Uhrhammer N, Pernin D, Presneau N, Tchirkov A, Vuillaume M, et al. High incidence of cancer in a family segregating a mutation of the ATM gene: possible role of ATM heterozygosity in cancer. Hum Mutat. 1999;14(6):485–92.
- Broeks A, de Klein A, Floore AN, Muijtjens M, Kleijer WJ, Jaspers NG, et al. ATM germline mutations in classical ataxia-telangiectasia patients in the Dutch population. Hum Mutat. 1998;12(5):330–7.
- Broeks A, Urbanus JH, Floore AN, Dahler EC, Klijn JG, Rutgers EJ, et al. ATM-heterozygous germline mutations contribute to breast cancer-susceptibility. Am J Hum Genet. 2000;66(2):494–500.
- Hsia T-C, Tsai C-W, Liang S-J, Chang W-S, Lin L-Y, Chen W-C, et al. Effects of ataxia telangiectasia mutated (ATM) genotypes and smoking habits on lung cancer risk in Taiwan. Anticancer Res. 2013;33(9):4067–71.
- Khalil H, Tummala H, Zhelev N. ATM in focus: A damage sensor and cancer target. Biodiscovery. Dundee Science Press; 2012;5.
- Li Z, Yu J, Zhang T, Li H, Ni Y. rs189037, a functional variant in ATM gene promoter, is associated with idiopathic nonobstructive azoospermia. Fertil Steril. Elsevier; 2013;100(6):1536–41.e1.
- Hadjisavvas A, Adamou A, O'Dowd Phanis C, Todd CM, Kitsios P, Kyriacou K, et al. Q356R and S1512I are BRCA1 variants that may be associated with breast cancer in a Cypriot family. Oncol Rep. 9(2):383–6.
- Spurdle AB, Hopper JL, Chen X, Dite GS, Cui J, McCredie MRE, et al. The BRCA2 372 HH genotype is associated with risk of breast cancer in Australian women under age 60 years. Cancer Epidemiol Biomarkers Prev. 2002;11(4):413–6.
- Healey CS, Dunning AM, Teare MD, Chase D, Parker L, Burn J, et al. A common variant in BRCA2 is associated with both breast cancer risk and prenatal viability. Nat Genet. 2000;26(3):362–4.
- Johnson N, Fletcher O, Palles C, Rudd M, Webb E, Sellick G, et al. Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Hum Mol Genet. 2007;16(9):1051–7.
- Wenham RM, Schildkraut JM, McLean K, Calingaert B, Bentley RC, Marks J, et al. Polymorphisms in BRCA1 and BRCA2 and risk of epithelial ovarian cancer. Clin Cancer Res. 2003;9(12):4396–403.
- Auranen A, Spurdle AB, Chen X, Lipscombe J, Purdie DM, Hopper JL, et al. BRCA2 Arg372Hispolymorphism and epithelial ovarian cancer risk. Int J Cancer. 2003;103(3):427–30.
- Figueiredo JC, Brooks JD, Conti D V, Poynter JN, Teraoka SN, Malone KE, et al. Risk of contralateral breast cancer associated with common variants in BRCA1 and BRCA2: potential modifying effect of BRCA1/BRCA2 mutation carrier status. Breast Cancer Res Treat. 2011;127(3):819–29.
- Valcárcel D, Martino R, Caballero D, Martin J, Ferra C, Nieto JB, et al. Sustained remissions of high-risk acute myeloid leukemia and myelodysplastic syndrome after reduced-intensity conditioning allogeneic hematopoietic transplantation: chronic graft-versus-host disease is the strongest factor improving survival. J Clin Oncol. 2008;26(4):577–84.
- Cho B-S, Lee S-E, Song H-H, Lee J-H, Yahng S-A, Eom K-S, et al. Graft-versus-tumor effect according to type of graft-versus-host disease defined by National Institutes of Health consensus criteria and associated outcomes. Biol Blood Marrow Transplant. 2012;18(7):1136–43.
- Rocha V, Cornish J, Sievers EL, Filipovich A, Locatelli F, Peters C, et al. Comparison of outcomes of unrelated bone marrow and umbilical cord blood transplants in children with acute leukemia. Blood. 2001;97(10):2962–71.
- Bhatia KG, Cherney BW, Huppi K, Magrath IT, Cossman J, Sausville E, et al. A Delection Linked to a Poly(ADP-ribose) Polymerase Gene on Chromosome 13q33-qter Occurs Frequently in the Normal Black Population as Well as in Multiple Tumor DNA. Cancer Res. 1990;50(17):5406–13.
- Lyn D, Cherney BW, Lalande M, Berenson JR, Lichtenstein A, Lupold S, et al. A duplicated region is responsible for the poly(ADP-ribose) polymerase polymorphism, on chromosome 13, associated with a predisposition to cancer. Am J Hum Genet. 1993;52(1):124–34.
- Doll J, Suarez B, Donis-Keller H. Association between prostate cancer in black Americans and an allele of the PADPRP pseudogene locus on chromosome 13. Am J Hum Genet. 1996;58(2):425–8.
- Enjuanes A, Benavente Y, Bosch F, Martín-Guerrero I, Colomer D, Pérez-Alvarez S, et al. Genetic variants in apoptosis and immunoregulation-related genes are associated with risk of chronic lymphocytic leukemia. Cancer Res. 2008;68(24):10178–86.
- Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92(11):874–97.
- Wu X, Zheng Y-L, Hsu TC. Mutagen-induced chromatid breakage as a marker of cancer risk. Methods Mol Biol. 2005;291:59–67.
- Rajaee-Behbahani N, Schmezer P, Risch A, Rittgen W, Kayser KW, Dienemann H, et al. Altered DNA repair capacity and bleomycin sensitivity as risk markers for non-small cell lung cancer. Int J Cancer. 2001;95(2):86–91.
- Zhu Y, Spitz MR, Zheng Y-L, Hong WK, Wu X. BPDE-induced lymphocytic 3p21.3 aberrations may predict head and neck carcinoma risk. Cancer. 2002;95(3):563–8.
- Wu X, Lippman SM, Lee JJ, Zhu Y, Wei QV, Thomas M, et al. Chromosome instability in lymphocytes: a potential indicator of predisposition to oral premalignant lesions. Cancer Res. 2002;62(10):2813–8.
- Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 310(5973):154–7.
- De Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, et al. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10(2):518–27.
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–14.
- Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247(4942):546–52.
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–5.
- Plunkett FJ, Soares M V, Annels N, Hislop A, Ivory K, Lowdell M, et al. The flow cytometric analysis of telomere length in antigen-specific CD8+ T cells during acute Epstein-Barr virus infection. Blood. 2001;97(3):700–7.
- Martens UM, Brass V, Sedlacek L, Pantic M, Exner C, Guo Y, et al. Telomere maintenance in human B lymphocytes. Br J Haematol. 2002;119(3):810–8.
- Lansdorp PM. Repair of telomeric DNA prior to replicative senescence. Mech Ageing Dev. 2000;118(1-2):23–34.
- Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295(5564):2446–9.
- Van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92(3):401–13.
- Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. Telomere shortening is proportional to the size of the G-rich telomeric 3'-overhang. J Biol Chem. 2000;275(26):19719–22.
- Keys B, Serra V, Saretzki G, Von Zglinicki T. Telomere shortening in human fibroblasts is not dependent on the size of the telomeric-3'-overhang. Aging Cell. 2004;3(3):103–9.
- Rahman R, Forsyth NR, Cui W. Telomeric 3'-overhang length is associated with the size of telomeres. Exp Gerontol. 2008;43(4):258–65.
- Calado RT, Regal JA, Kleiner DE, Schrump DS, Peterson NR, Pons V, et al. A spectrum of severe familial liver disorders associate with telomerase mutations. PLoS One. 2009;4(11):e7926.
- Betjes MGH, Langerak AW, van der Spek A, de Wit EA, Litjens NHR. Premature aging of circulating T cells in patients with end-stage renal disease. Kidney Int. 2011;80(2):208–17.
- Cesselli D, Beltrami AP, D'Aurizio F, Marcon P, Bergamin N, Toffoletto B, et al. Effects of age and heart failure on human cardiac stem cell function. Am J Pathol. 2011;179(1):349–66.
- Costenbader KH, Prescott J, Zee RY, De Vivo I. Immunosenescence and rheumatoid arthritis: does telomere shortening predict impending disease? Autoimmun Rev. 2011;10(9):569–73.
- Noureddine H, Gary-Bobo G, Alifano M, Marcos E, Saker M, Vienney N, et al. Pulmonary artery smooth muscle cell senescence is a pathogenic mechanism for pulmonary hypertension in chronic lung disease. Circ Res. 2011;109(5):543–53.
- Ilmonen P, Kotrschal A, Penn DJ. Telomere attrition due to infection. Masucci MG, editor. PLoS One. Public Library of Science; 2008;3(5):e2143.
- Kotrschal A, Ilmonen P, Penn DJ. Stress impacts telomere dynamics. Biol Lett. 2007;3(2):128–30.
- Kananen L, Surakka I, Pirkola S, Suvisaari J, Lönnqvist J, Peltonen L, et al. Childhood adversities are associated with shorter telomere length at adult age both in individuals with an anxiety disorder and controls. Mitchell AJ, editor. PLoS One. Public Library of Science; 2010;5(5):e10826.
- Drury SS, Theall K, Gleason MM, Smyke AT, De Vivo I, Wong JYY, et al. Telomere length and early severe social deprivation: linking early adversity and cellular aging. Mol Psychiatry. 2012;17(7):719–27.
- Parks CG, DeRoo LA, Miller DB, McCanlies EC, Cawthon RM, Sandler DP. Employment and work schedule are related to telomere length in women. Occup Environ Med. 2011;68(8):582–9.
- Aviv A, Chen W, Gardner JP, Kimura M, Brimacombe M, Cao X, et al. Leukocyte telomere dynamics: longitudinal findings among young adults in the Bogalusa Heart Study. Am J Epidemiol. 2009;169(3):323–9.
- Svenson U, Nordfjäll K, Baird D, Roger L, Osterman P, Hellenius M-L, et al. Blood cell telomere length is a dynamic feature. Cotterill S, editor. PLoS One. Public Library of Science; 2011;6(6):e21485.
- Atanassov BS, Ninova PD, Anachkova BB, Russev GC. Relationship between DNA repair capacity and resistance to genotoxins in four human cell lines. Cancer Detect Prev. 2003;27(1):24–9.
- Furgason JM, Bahassi EM. Targeting DNA repair mechanisms in cancer. Pharmacol Ther. 2013;137(3):298–308.
- Moghaddam AA, Woodward M, Huxley R. Obesity and risk of colorectal cancer: a meta-analysis of 31 studies with 70,000 events. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2533–47.
- Hamilton M, Wolf JL, Rusk J, Beard SE, Clark GM, Witt K, et al. Effects of smoking on the pharmacokinetics of erlotinib. Clin Cancer Res. 2006;12(7 Pt 1):2166–71.
- Rocha-Lima CM, Raez LE. Erlotinib (tarceva) for the treatment of non-small-cell lung cancer and pancreatic cancer. P T. 2009;34(10):554–64.
- Chen J-Q, Russo J. ERalpha-negative and triple negative breast cancer: molecular features and potential therapeutic approaches. Biochim Biophys Acta. 2009;1796(2):162–75.
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–10.
- Oh E-T, Park M-T, Choi B-H, Ro S, Choi E-K, Jeong S-Y, et al. Novel histone deacetylase inhibitor CG200745 induces clonogenic cell death by modulating acetylation of p53 in cancer cells. Invest New Drugs. 2012;30(2):435–42.
- Huang C. Resveratrol suppresses cell transformation and induces apoptosis through a p53-dependent pathway. Carcinogenesis. 1999;20(2):237–42.
- She QB, Bode AM, Ma WY, Chen NY, Dong Z. Resveratrol-induced activation of p53 and apoptosis is mediated by extracellular-signal-regulated protein kinases and p38 kinase. Cancer Res. 2001;61(4):1604–10.
- Tovar C, Graves B, Packman K, Filipovic Z, Higgins B, Xia M, et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73(8):2587–97.
- Zenz T, Benner A, Döhner H, Stilgenbauer S. Chronic lymphocytic leukemia and treatment resistance in cancer: the role of the p53 pathway. Cell Cycle. 2008;7(24):3810–4.
- Cobo M, Isla D, Massuti B, Montes A, Sanchez JM, Provencio M, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25(19):2747–54.
- Fujita H, Ohuchida K, Mizumoto K, Itaba S, Ito T, Nakata K, et al. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12(10):807–17.
- Nakamura J, Kohya N, Kai K, Ohtaka K, Hashiguchi K, Hiraki M, et al. Ribonucleotide reductase subunit M1 assessed by quantitative double-fluorescence immunohistochemistry predicts the efficacy of gemcitabine in biliary tract carcinoma. Int J Oncol. 2010;37(4):845–52.
- Wang L, Zhang G, Chen J, Li J, Li M, Xu N, et al. RRM1 gene expression in peripheral blood is predictive of shorter survival in Chinese patients with advanced non-small-cell lung cancer treated by gemcitabine and platinum. J Zhejiang Univ Sci B. 2011;12(3):174–9.
- Zeng H, Yu H, Lu L, Jain D, Kidd MS, Saif MW, et al. Genetic effects and modifiers of radiotherapy and chemotherapy on survival in pancreatic cancer. Pancreas. 2011;40(5):657–63.
- Park DJ, Zhang W, Stoehlmacher J, Tsao-Wei D, Groshen S, Gil J, et al. ERCC1 gene polymorphism as a predictor for clinical outcome in advanced colorectal cancer patients treated with platinum-based chemotherapy. Clin Adv Hematol Oncol. 2003;1(3):162–6.
- Zhou W, Gurubhagavatula S, Liu G, Park S, Neuberg DS, Wain JC, et al. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2004;10(15):4939–43.
- Wu X, Lu C, Ye Y, Chang J, Yang H, Lin J, et al. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharmacogenet Genomics. 2008;18(11):955–65.
- Drach J, Ackermann J, Fritz E, Krömer E, Schuster R, Gisslinger H, et al. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood. 1998;92(3):802–9.
- Bansal A, Soni A, Rao P, Singh LC, Mishra AK, Mohanty NK, et al. Implication of DNA repair genes in prostate tumourigenesis in Indian males. Indian J Med Res. 2012;136(4):622–32.
- Ohnstad HO, Castro R, Sun J, Heintz K-M, Vassilev LT, Bjerkehagen B, et al. Correlation of TP53 and MDM2 genotypes with response to therapy in sarcoma. Cancer. 2013;119(5):1013–22.
- Bonafé M, Ceccarelli C, Farabegoli F, Santini D, Taffurelli M, Barbi C, et al. Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res. 2003;9(13):4860–4.
- Boldrini L, Gisfredi S, Ursino S, Lucchi M, Greco G, Mussi A, et al. Effect of the p53 codon 72 and intron 3 polymorphisms on non-small cell lung cancer (NSCLC) prognosis. Cancer Invest. 2008;26(2):168–72.
- Wu W, Li H, Wang H, Zhao X, Gao Z, Qiao R, et al. Effect of polymorphisms in XPD on clinical outcomes of platinum-based chemotherapy for Chinese non-small cell lung cancer patients. PLoS One. 2012;7(3):e33200.
- Qiu M, Yang X, Hu J, Ding X, Jiang F, Yin R, et al. Predictive value of XPD polymorphisms on platinum-based chemotherapy in non-small cell lung cancer: a systematic review and meta-analysis. Miao X, editor. PLoS One. Public Library of Science; 2013;8(8):e72251.
- Zhang L, Ma W, Li Y, Wu J, Shi GY. Pharmacogenetics of DNA repair gene polymorphisms in non-small-cell lung carcinoma patients on platinum-based chemotherapy. Genet Mol Res. 2014;13(1):228–36.
- Strom SS, Estey E, Outschoorn UM, Garcia-Manero G. Acute myeloid leukemia outcome: role of nucleotide excision repair polymorphisms in intermediate risk patients. Leuk Lymphoma. 2010;51(4):598–605.
- Fröhling S, Schlenk RF, Kayser S, Morhardt M, Benner A, Döhner K, et al. Cytogenetics and age are major determinants of outcome in intensively treated acute myeloid leukemia patients older than 60 years: results from AMLSG trial AML HD98-B. Blood. 2006;108(10):3280–8.
- Grimwade D, Hills RK, Moorman A V, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–65.
- Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, et al. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22(13):2594–601.
- Booton R, Ward T, Heighway J, Taylor P, Power F, Ashcroft L, et al. Xeroderma pigmentosum group D haplotype predicts for response, survival, and toxicity after platinum-based chemotherapy in advanced nonsmall cell lung cancer. Cancer. 2006;106(11):2421–7.
- Metzger R, Warnecke-Eberz U, Alakus H, Kütting F, Brabender J, Vallböhmer D, et al. Neoadjuvant radiochemotherapy in adenocarcinoma of the esophagus: ERCC1 gene polymorphisms for prediction of response and prognosis. J Gastrointest Surg. 2012;16(1):26–34; discussion 34.
- Matakidou A, el Galta R, Webb EL, Rudd MF, Bridle H, Eisen T, et al. Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet. 2007;16(19):2333–40.
- Olaussen KA, Dunant A, Fouret P, Brambilla E, André F, Haddad V, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355(10):983–91.
- Bauman JE, Austin MC, Schmidt R, Kurland BF, Vaezi A, Hayes DN, et al. ERCC1 is a prognostic biomarker in locally advanced head and neck cancer: results from a randomised, phase II trial. Br J Cancer. 2013;109(8):2096–105.
- Chen C, Wang F, Wang Z, Li C, Luo H, Liang Y, et al. Polymorphisms in ERCC1 C8092A predict progression-free survival in metastatic/recurrent nasopharyngeal carcinoma treated with cisplatin-based chemotherapy. Cancer Chemother Pharmacol. 2013;72(2):315–22.
- Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, et al. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10(2):118–23.
- Moxley KM, Benbrook DM, Queimado L, Zuna RE, Thompson D, McCumber M, et al. The role of single nucleotide polymorphisms of the ERCC1 and MMS19 genes in predicting platinum-sensitivity, progression-free and overall survival in advanced epithelial ovarian cancer. Gynecol Oncol. 2013;130(2):377–82.
- Ryu J-S, Hong Y-C, Han H-S, Lee J-E, Kim S, Park Y-M, et al. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44(3):311–6.
- Isla D, Sarries C, Rosell R, Alonso G, Domine M, Taron M, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15(8):1194–203.
- Kamikozuru H, Kuramochi H, Hayashi K, Nakajima G, Yamamoto M. ERCC1 codon 118 polymorphism is a useful prognostic marker in patients with pancreatic cancer treated with platinum-based chemotherapy. Int J Oncol. 2008;32(5):1091–6.
- Sun X, Li F, Sun N, Shukui Q, Baoan C, Jifeng F, et al. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009;65(2):230–6.
- Khalil HS, Tummala H, Oluwaseun OA, Zhelev N. Novel insights of Ataxia Telangiectasia Mutated (ATM) regulation and its potential as a target for therapeutic intervention in cancer. Curr Opin Biotechnol. 2011;22:S115–S116.
- Khalil HS, Tummala H, Hupp TR, Zhelev N. Pharmacological inhibition of ATM by KU55933 stimulates ATM transcription. Exp Biol Med (Maywood). 2012;237(6):622–34.
- Idowu M. Reverse engineering of drug induced DNA damage response signalling pathway reveals dual outcomes of ATM kinase inhibition. Biodiscovery. Dundee Science Press; 2013;4.
- Whittaker S, Walton M, Kelland L, Garrett M, Zhelev N, Workman P. Rb Phorphorylation as a pharmacodynamic marker of Roscovitine (CYC202) activity in vitro and in vivo. Proc Am Assoc Cancer Res. 2001;42:926.
- McClue SJ, Blake D, Clarke R, Cowan A, Cummings L, Fischer PM, et al. In vitro and in vivo antitumor properties of the cyclin dependent kinase inhibitor CYC202 (R-roscovitine). Int J Cancer. 2002;102(5):463–8.
- Zhang G-J, Safran M, Wei W, Sorensen E, Lassota P, Zhelev N, et al. Bioluminescent imaging of Cdk2 inhibition in vivo. Nat Med. 2004;10(6):643–8.
- Atanasova G, Jans R, Zhelev N, Mitev V, Poumay Y. Effects of the cyclin-dependent kinase inhibitor CYC202 (R-roscovitine) on the physiology of cultured human keratinocytes. Biochem Pharmacol. 2005;70(6):824–36.
- Zhelev N, Tummala H, Trifonov D, D'Ascanio I, Adebola Oluwaseun O, Fischer PM. Recent advances in the development of cyclin-dependent kinase inhibitors as new therapeutics in oncology and cardiology. Curr Opin Biotechnol. 2013;24:S25.
- Trifonov D, Tummala H, Clements S, Zhelev N. Effect of roscovitine on cardiac hypertrophy in human stem cell derived cardiomyocytes. Curr Opin Biotechnol. 2013;24:S114–S115.
- Rosell R, Taron M, Camps C, López-Vivanco G. Influence of genetic markers on survival in non-small cell lung cancer. Drugs Today (Barc). 2003;39(10):775–86.
- Le May N, Egly J-M, Coin F. True lies: the double life of the nucleotide excision repair factors in transcription and DNA repair. J Nucleic Acids. 2010;2010.
- Quintela-Fandino M, Hitt R, Medina PP, Gamarra S, Manso L, Cortes-Funes H, et al. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24(26):4333–9.
- Wang Y, Chen J, Li X, He Y, Hu B, Ji C, et al. Genetic polymorphisms of ERCC1 and their effects on the efficacy of cisplatin-based chemotherapy in advanced esophageal carcinoma. Oncol Rep. 2011;25(4):1047–52.
- Takenaka T, Yano T, Kiyohara C, Miura N, Kouso H, Ohba T, et al. Effects of excision repair cross-complementation group 1 (ERCC1) single nucleotide polymorphisms on the prognosis of non-small cell lung cancer patients. Lung Cancer. 2010;67(1):101–7.
- Elnenaei M, Gruszka-Westwood A, A'Hernt R, Matutes E, Sirohi B, Powles R, et al. Gene abnormalities in multiple myeloma; the relevance of TP53, MDM2, and CDKN2A. Haematologica. 2003;88(5):529–37.
- Twardella D, Popanda O, Helmbold I, Ebbeler R, Benner A, von Fournier D, et al. Personal characteristics, therapy modalities and individual DNA repair capacity as predictive factors of acute skin toxicity in an unselected cohort of breast cancer patients receiving radiotherapy. Radiother Oncol. 2003;69(2):145–53.
- Rogers PC, Meacham LR, Oeffinger KC, Henry DW, Lange BJ. Obesity in pediatric oncology. Pediatr Blood Cancer. 2005;45(7):881–91.
- Hakimi AA, Furberg H, Zabor EC, Jacobsen A, Schultz N, Ciriello G, et al. An epidemiologic and genomic investigation into the obesity paradox in renal cell carcinoma. J Natl Cancer Inst. 2013;105(24):1862–70.
- Petkova R, Chelenkova P, Georgieva E, Chakarov S. What's your poison? Impact of individual repair capacity on the outcomes of genotoxic therapies in cancer. Part II - information content and validity of biomarkers for individual repair capacity in the assessment of outcomes of anticancer therapy. Biotechnol Biotechnol Eq. 06 May 2014 DOI:10.1080/13102818.2014.902532),
- Chang-Claude J, Popanda O, Tan X-L, Kropp S, Helmbold I, von Fournier D, et al. Association between polymorphisms in the DNA repair genes, XRCC1, APE1, and XPD and acute side effects of radiotherapy in breast cancer patients. Clin Cancer Res. 2005;11(13):4802–9.
- Wu W, Zhang W, Qiao R, Chen D, Wang H, Wang Y, et al. Association of XPD polymorphisms with severe toxicity in non-small cell lung cancer patients in a Chinese population. Clin Cancer Res. 2009;15(11):3889–95.
- Sakano S, Hinoda Y, Sasaki M, Wada T, Matsumoto H, Eguchi S, et al. Nucleotide excision repair gene polymorphisms may predict acute toxicity in patients treated with chemoradiotherapy for bladder cancer. Pharmacogenomics. 2010;11(10):1377–87.
- Damaraju S, Murray D, Dufour J, Carandang D, Myrehaug S, Fallone G, et al. Association of DNA repair and steroid metabolism gene polymorphisms with clinical late toxicity in patients treated with conformal radiotherapy for prostate cancer. Clin Cancer Res. 2006;12(8):2545–54.
- Yoon HH, Catalano P, Gibson MK, Skaar TC, Philips S, Montgomery EA, et al. Genetic variation in radiation and platinum pathways predicts severe acute radiation toxicity in patients with esophageal adenocarcinoma treated with cisplatin-based preoperative radiochemotherapy: results from the Eastern Cooperative Oncology Group. Cancer Chemother Pharmacol. 2011;68(4):863–70.
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1(6):783–93.
- Bartsch H, Dally H, Popanda O, Risch A, Schmezer P. Genetic risk profiles for cancer susceptibility and therapy response. Recent Results Cancer Res. 2007;174:19–36.
- Santi R, Cetica V, Franchi A, Pepi M, Cesinaro AM, Miracco C, et al. Tumour suppressor gene TP53 mutations in atypical vascular lesions of breast skin following radiotherapy. Histopathology. 2011;58(3):455–66.
- Angèle S, Romestaing P, Moullan N, Vuillaume M, Chapot B, Friesen M, et al. ATM haplotypes and cellular response to DNA damage: association with breast cancer risk and clinical radiosensitivity. Cancer Res. 2003;63(24):8717–25.
- Zhang L, Yang M, Bi N, Fang M, Sun T, Ji W, et al. ATM polymorphisms are associated with risk of radiation-induced pneumonitis. Int J Radiat Oncol Biol Phys. 2010;77(5):1360–8.
- Xiong H, Liao Z, Liu Z, Xu T, Wang Q, Liu H, et al. ATM polymorphisms predict severe radiation pneumonitis in patients with non-small cell lung cancer treated with definitive radiation therapy. Int J Radiat Oncol Biol Phys. 2013;85(4):1066–73.
- Suk R, Gurubhagavatula S, Park S, Zhou W, Su L, Lynch TJ, et al. Polymorphisms in ERCC1 and grade 3 or 4 toxicity in non-small cell lung cancer patients. Clin Cancer Res. 2005;11(4):1534–8.
- Yu T, Liu Y, Lu X, Xiao S, Cai Y, Jin C, et al. Excision repair of BPDE-adducts in human lymphocytes: diminished capacity associated with ERCC1 C8092A (rs3212986) polymorphism. Arch Toxicol. 2013;87(4):699–709.
- Pérez-Soler R, Delord JP, Halpern A, Kelly K, Krueger J, Sureda BM, et al. HER1/EGFR inhibitor-associated rash: future directions for management and investigation outcomes from the HER1/EGFR inhibitor rash management forum. Oncologist. 2005;10(5):345–56.
- Dienstmann R, Braña I, Rodon J, Tabernero J. Toxicity as a biomarker of efficacy of molecular targeted therapies: focus on EGFR and VEGF inhibiting anticancer drugs. Oncologist. 2011;16(12):1729–40.
- Kane RC, Farrell AT, Saber H, Tang S, Williams G, Jee JM, et al. Sorafenib for the treatment of advanced renal cell carcinoma. Clin Cancer Res. 2006;12(24):7271–8.
- Motzer RJ, Rini BI, Bukowski RM, Curti BD, George DJ, Hudes GR, et al. Sunitinib in patients with metastatic renal cell carcinoma. JAMA. 2006;295(21):2516–24.
- An MM, Zou Z, Shen H, Liu P, Chen ML, Cao YB, et al. Incidence and risk of significantly raised blood pressure in cancer patients treated with bevacizumab: an updated meta-analysis. Eur J Clin Pharmacol. 2010;66(8):813–21.
- Scartozzi M, Galizia E, Chiorrini S, Giampieri R, Berardi R, Pierantoni C, et al. Arterial hypertension correlates with clinical outcome in colorectal cancer patients treated with first-line bevacizumab. Ann Oncol. 2009;20(2):227–30.
- Bono P, Elfving H, Utriainen T, Osterlund P, Saarto T, Alanko T, et al. Hypertension and clinical benefit of bevacizumab in the treatment of advanced renal cell carcinoma. Ann Oncol. 2009;20(2):393–4.
- Kim JJ, Vaziri SAJ, Rini BI, Elson P, Garcia JA, Wirka R, et al. Association of VEGF and VEGFR2 single nucleotide polymorphisms with hypertension and clinical outcome in metastatic clear cell renal cell carcinoma patients treated with sunitinib. Cancer. 2012;118(7):1946–54.
- Wolter P, Stefan C, Decallonne B, Dumez H, Bex M, Carmeliet P, et al. The clinical implications of sunitinib-induced hypothyroidism: a prospective evaluation. Br J Cancer. 2008;99(3):448–54.
- Schmidinger M, Vogl UM, Bojic M, Lamm W, Heinzl H, Haitel A, et al. Hypothyroidism in patients with renal cell carcinoma: blessing or curse? Cancer. 2011;117(3):534–44.
- Hsu TC, Johnston DA, Cherry LM, Ramkissoon D, Schantz SP, Jessup JM, et al. Sensitivity to genotoxic effects of bleomycin in humans: possible relationship to environmental carcinogenesis. Int J Cancer. 1989;43(3):403–9.
- Li C, Wang L-E, Wei Q. DNA repair phenotype and cancer susceptibility--a mini review. Int J Cancer. 2009;124(5):999–1007.
- Abdel-Rahman SZ, El-Zein RA. Evaluating the effects of genetic variants of DNA repair genes using cytogenetic mutagen sensitivity approaches. Biomarkers. 2011;16(5):393–404.
- Kohn KW, Grimek-Ewig RA. Alkaline Elution Analysis, a New Approach to the Study of DNA Single-Strand Interruptions in Cells. Cancer Res. 1973;33(8):1849–53.
- Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486(4):217–47.
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40(2):359–69.
- Cedervall B, Wong R, Albright N, Dynlacht J, Lambin P, Dewey WC. Methods for the quantification of DNA double-strand breaks determined from the distribution of DNA fragment sizes measured by pulsed-field gel electrophoresis. Radiat Res. 1995;143(1):8–16.
- Bryant HE. DNA double-strand break damage and repair assessed by pulsed-field gel electrophoresis. Methods Mol Biol. 2012;920:315–21.
- Herschleb J, Ananiev G, Schwartz DC. Pulsed-field gel electrophoresis. Nat Protoc. 2007;2(3):677–84.
- Vrána O, Boudný V, Brabec V. Superhelical torsion controls DNA interstrand cross-linking by antitumor cis- diamminedichloroplatinum(II). Nucleic Acids Res. 1996;24(20):3918–25.
- Thompson RJ, Mosig G. Light affects the structure of Chlamydomonas chloroplast chromosomes. Nucleic Acids Res. 1990;18(9):2625–31.
- Dresler W, Stein R. Ueber den Hydroxyharnstoff. Justus Liebigs Ann Chem Pharmacol. 1869;150:242–52.
- Nutter L, Cheng Y. Nature and properties of mammalian ribonucleoside diphosphate reductase. In: Cory J, Cory A, editors. Inhibitors of ribonucleoside diphosphate reductase activity. New York: Pergamon Press; 1989. p. 37.
- Donehower RC. Hydroxyurea. In: Chabner B, Collins J, editors. Cancer chemotherapy: principles and practice. Philadelphia: Lippincott; 1990. p. 154–79.
- Timson J. Hydroxyurea. Mutat Res. 1975;32(2):115–32.
- Chakarov S, Stoilov P, Alexandrov A, Russev G. Repair pattern in the beta-globin gene cluster of human fibroblasts after ultraviolet irradiation. Eur J Biochem. 1997;248(3):669–75.
- Chakalova L, Russev G. Transcriptionally active and inactive mouse beta-globin gene loci are repaired at similar rates after ultraviolet irradiation. Eur J Biochem. 1999;261(3):667–73.
- Marden A, Walmsley RM, Schweizer LM, Schweizer M. Yeast-based assay for the measurement of positive and negative influences on microsatellite stability. FEMS Yeast Res. 2006;6(5):716–25.
- Chakarov S, Roeva I, Russev G. An Experimental Model for Assessment of Global DNA Repair Capacity. Biotechnol Biotechnol Eq. 2011;25(3):2505–7.
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239(4839):487–91.
- Kalinowski DP, Illenye S, Van Houten B. Analysis of DNA damage and repair in murine leukemia L1210 cells using a quantitative polymerase chain reaction assay. Nucleic Acids Res. 1992;20(13):3485–94.
- Ploskonosova II, Baranov VI, Gaziev AI. Estimation of DNA damage and repair in tissues of gamma-irradiated animals using the polymerase chain reaction. Biochem Biokhimiia. 1999;64(11):1320–5.
- Zhang N, Lu X, Legerski RJ. Partial reconstitution of human interstrand cross-link repair in vitro: characterization of the roles of RPA and PCNA. Biochem Biophys Res Commun. 2003;309(1):71–8.
- Gospodinov A, Russev G, Anachkova B. Determination of DNA repair capacity of protein extracts by restoration of the transformation efficiency of damaged plasmids. Anal Biochem. 2003;315(2):285–8.
- Gospodinov A, Anachkova B. Cells synchronized in S phase show increased rate of repair of UV damaged plasmids. FEBS Lett. 2004;572(1-3):99–102.
- Hochleitner K, Thomale J, Nikitin AYu, Rajewsky MF. Monoclonal antibody-based, selective isolation of DNA fragments containing an alkylated base to be quantified in defined gene sequences. Nucleic Acids Res. 1991;19(16):4467–72.
- Kobayashi N, Katsumi S, Imoto K, Nakagawa A, Miyagawa S, Furumura M, et al. Quantitation and visualization of ultraviolet-induced DNA damage using specific antibodies: application to pigment cell biology. Pigment Cell Res. 2001;14(2):94–102.
- Cooke MS, Robson A. Immunochemical detection of UV-induced DNA damage and repair. Methods Mol Biol. 2006;314:215–28.
- Al-Adhami BH, Nichols RAB, Kusel JR, O'Grady J, Smith H V. Detection of UV-induced thymine dimers in individual Cryptosporidium parvum and Cryptosporidium hominis oocysts by immunofluorescence microscopy. Appl Environ Microbiol. 2007;73(3):947–55.
- Kuo LJ, Yang L-X. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 22(3):305–9.
- Nakamura AJ, Rao VA, Pommier Y, Bonner WM. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle. 2010;9(2):389–97.
- Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos Trans R Soc Lond B Biol Sci. 2004;359(1441):87–93.
- Balajee AS, Dianova I, Bohr VA. Oxidative damage-induced PCNA complex formation is efficient in xeroderma pigmentosum group A but reduced in Cockayne syndrome group B cells. Nucleic Acids Res. 1999;27(22):4476–82.
- Hildrestrand GA, Rolseth V, Bjørås M, Luna L. Human NEIL1 localizes with the centrosomes and condensed chromosomes during mitosis. DNA Repair (Amst). 2007;6(10):1425–33.
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283(39):26349–56.
- Cadet J, Douki T, Frelon S, Sauvaigo S, Pouget J-P, Ravanat J-L. Assessment of oxidative base damage to isolated and cellular DNA by HPLC-MS/MS measurement. Free Radic Biol Med. 2002;33(4):441–9.
- Danielsen PH, Bräuner EV, Barregard L, Sällsten G, Wallin M, Olinski R, et al. Oxidatively damaged DNA and its repair after experimental exposure to wood smoke in healthy humans. Mutat Res. 2008;642(1-2):37–42.
- Jaruga P, Kirkali G, Dizdaroglu M. Measurement of formamidopyrimidines in DNA. Free Radic Biol Med. 2008;45(12):1601–9.
- Chan W, Chen B, Wang L, Taghizadeh K, Demott MS, Dedon PC. Quantification of the 2-deoxyribonolactone and nucleoside 5'-aldehyde products of 2-deoxyribose oxidation in DNA and cells by isotope-dilution gas chromatography mass spectrometry: differential effects of gamma-radiation and Fe2+-EDTA. J Am Chem Soc. 2010;132(17):6145–53.
- Pham P, Zhang K, Goodman MF. Hypermutation at A/T sites during G.U mismatch repair in vitro by human B-cell lysates. J Biol Chem. 2008;283(46):31754–62.
- Schanz S, Castor D, Fischer F, Jiricny J. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci U S A. 2009;106(14):5593–8.
- Sharma S, Raghavan SC. Nonhomologous DNA end joining in cell-free extracts. J Nucleic Acids. 2010;2010.
- Protić-Sabljić M, Kraemer KH. Host cell reactivation by human cells of DNA expression vectors damaged by ultraviolet radiation or by acid-heat treatment. Carcinogenesis. 1986;7(10):1765–70.
- Henderson EE, Valerie K, Green AP, de Riel JK. Host cell reactivation of CAT-expression vectors as a method to assay for cloned DNA-repair genes. Mutat Res. 220(2-3):151–60.
- Colicos MA, Haj-Ahmad Y, Valerie K, Henderson EE, Rainbow AJ. Construction of a recombinant adenovirus containing the denV gene from bacteriophage T4 which can partially restore the DNA repair deficiency in xeroderma pigmentosum fibroblasts. Carcinogenesis. 1991;12(2):249–55.
- Athas WF, Hedayati MA, Matanoski GM, Farmer ER, Grossman L. Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res. 1991;51(21):5786–93.
- Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, et al. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res. 2002;509(1-2):165–74.
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9(6):675–82.
- Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst). 2007;6(6):781–8.
- Wang L, Wei Q, Shi Q, Guo Z, Qiao Y, Spitz MR. A modified host-cell reactivation assay to measure repair of alkylating DNA damage for assessing risk of lung adenocarcinoma. Carcinogenesis. 2007;28(7):1430–6.
- Landi MT, Baccarelli A, Tarone RE, Pesatori A, Tucker MA, Hedayati M, et al. DNA repair, dysplastic nevi, and sunlight sensitivity in the development of cutaneous malignant melanoma. J Natl Cancer Inst. 2002;94(2):94–101.
- Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2003;12(8):689–98.
- Collins AR. Measuring oxidative damage to DNA and its repair with the comet assay. Biochim Biophys Acta. 2014;1840(2):794–800.
- Thomas EA, Thomas CA. Nucleoid halo expansion indirectly measures DNA damage in single cells. Exp Cell Res. 1989;183(1):149–58.
- Fortini P, Raspaglio G, Falchi M, Dogliotti E. Analysis of DNA alkylation damage and repair in mammalian cells by the comet assay. Mutagenesis. 1996;11(2):169–75.
- Møller P. The alkaline comet assay: towards validation in biomonitoring of DNA damaging exposures. Basic Clin Pharmacol Toxicol. 2006;98(4):336–45.
- Merk O, Speit G. Detection of crosslinks with the comet assay in relationship to genotoxicity and cytotoxicity. Environ Mol Mutagen. 1999;33(2):167–72.
- Wynne P, Newton C, Ledermann JA, Olaitan A, Mould TA, Hartley JA. Enhanced repair of DNA interstrand crosslinking in ovarian cancer cells from patients following treatment with platinum-based chemotherapy. Br J Cancer. 2007;97(7):927–33.
- Angelis KJ, Dusinská M, Collins AR. Single cell gel electrophoresis: detection of DNA damage at different levels of sensitivity. Electrophoresis. 1999;20(10):2133–8.
- Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, et al. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35(3):206–21.
- Azqueta A, Lorenzo Y, Collins AR. In vitro comet assay for DNA repair: a warning concerning application to cultured cells. Mutagenesis. 2009;24(4):379–81.
- Collins AR. The comet assay. Principles, applications, and limitations. Methods Mol Biol. 2002;203:163–77.
- Spyridopoulos I, Erben Y, Brummendorf TH, Haendeler J, Dietz K, Seeger F, et al. Telomere gap between granulocytes and lymphocytes is a determinant for hematopoetic progenitor cell impairment in patients with previous myocardial infarction. Arterioscler Thromb Vasc Biol. 2008;28(5):968–74.
- Petkova R, Chicheva Z, Chakarov S. Measuring Telomere Length – From Ends To Means. Biotechnol Biotechnol Eq. 2011;25(4):2576–82.
- Allshire RC, Dempster M, Hastie ND. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 1989;17(12):4611–27.
- Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47.
- Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37(3):e21.
- Baerlocher GM, Mak J, Tien T, Lansdorp PM. Telomere length measurement by fluorescence in situ hybridization and flow cytometry: tips and pitfalls. Cytometry. 2002;47(2):89–99.
- Kapoor V, Telford WG. Telomere length measurement by fluorescence in situ hybridization and flow cytometry. Methods Mol Biol. 2004;263:385–98.
- Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365(6446):566–8.
- Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc. Nature Publishing Group; 2006;1(5):2365–76.
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33(2):203–7.
- Hills M, Lücke K, Chavez EA, Eaves CJ, Lansdorp PM. Probing the mitotic history and developmental stage of hematopoietic cells using single telomere length analysis (STELA). Blood. 2009;113(23):5765–75.
- Piatyszek MA, Kim NW, Weinrich SL, Hiyama K, Hiyama E, Wright WE, et al. Detection of telomerase activity in human cells and tumors by a telomeric repeat amplification protocol (TRAP). Methods Cell Sci. 1995;17(1):1–15.
- Hiyama E, Yokoyama T, Tatsumoto N, Hiyama K, Imamura Y, Murakami Y, et al. Telomerase activity in gastric cancer. Cancer Res. 1995;55(15):3258–62.
- Fajkus J. Detection of telomerase activity by the TRAP assay and its variants and alternatives. Clin Chim Acta. 2006;371(1-2):25–31.
- Hansen K, Sickelmann F, Pietrowsky R, Fehm HL, Born J. Systemic immune changes following meal intake in humans. Am J Physiol. 1997;273(2 Pt 2):R548–53.