1. Cooking up cancer – how from a tiny DNA alteration may eventually grow a large tumour
Torchwood: Miracle Day's official tagline (2011)
Systematic and efficient repair of DNA damage, implemented as soon as possible after the damage has been detected, is the main mechanism of protection of living cells against potentially harmful modifications in their major information carrier molecule. The DNA repair system of the average individual (apart from the relatively rare exceptions of inherited DNA repair deficiencies), coupled with the mechanism of programmed cell death, function together well enough so as not to allow the occurrence of too many alterations of the genetic content per cell, or, at least, that they would not be passed on to the cell's progeny. There are, however, sources of DNA errors that cannot be eliminated or avoided. As it was already discussed, despite the accuracy of template copying during replication and DNA repair, the underlying mechanism is essentially error-prone. The risk of occurrence of a copying error and the associated risk of the error becoming fixed heritable mutation are minute between two successive cellular generations, but tend to accumulate over time. As the cell and the organism age, the efficiency of the mechanisms for detection and repair of DNA damage generally declines. This is associated with accumulation of unrepaired damage in DNA and, respectively, with increased risk of introduction of mutations. The mutations may affect various genes, but pro-carcinogenic action of random mutagenesis is usually most pronounced when directly affecting genes coding for products that are associated with stimulation of cell proliferation (e.g. growth factors and their receptors) and/or products that may suppress cell growth in response to damage-associated signalling (e.g. TP53, ATM, BRCA1 and 2, CHK proteins, etc.). Some of the mutations that occur de novo in somatic cells may produce a net increase in the genome mutability, which may, in turn, result in higher rate of mutation occurrence per cell generation. Other somatic mutations may result in inactivation or evasion of one or more pathways and mechanisms for detection and/or repair of DNA damage, or the mechanisms that dictate that a damaged cell must die. Thus, the minute error/s occurring in every cell division may, with time, multiply and augment each other. Eventually, this may result in cell transformation and, later, in overt cancer. Usually this occurs years and decades after the initial event/s that supposedly triggered the process.
It would be safe to say that high-fidelity copying of DNA during replication, together with the concerted action of the mechanisms of DNA repair and programmed cell death are normally sufficient to sustain the individual healthy from conception up to middle adulthood (45–55 years). Beyond that age, the risk of occurrence of somatic mutation/s eventually resulting in cancer may become significant. Despite what the media may say, cancer is (and always was) predominantly a disease of middle and advanced age. Not all people beyond the age of 60, however, eventually develop cancer, and many of the oldest old (>85) remain cancer-free to their deaths.
The rate of accumulation of mutations is usually dependent of the number of divisions the cell goes through. Different cell types have different turnover rates. Cell types with naturally rapid turnover rate are, for example, neutrophil granulocytes (replaced about every 4–5 days); cells in the intestinal crypts (about every 7 days), and erythrocytes (about every 120 days). Adipocytes are exemplary slow dividers, being replaced at a rate of 6–8% per year. Some terminally differentiated cells that were initially believed to be irreplaceable, such as neurons in the olfactory bulb in mammals, are actually replaced about every 6 weeks [
There are four basic pathways by which a normal cell may be transformed to a cancer cell. The origins of all may be traced to instances of DNA damage becoming fixed as permanent alterations in the sequence and/or the structure of DNA.
- Abrogating the restrictions normally placed upon cell division, resulting in increased proliferative capacity of the cell (e.g. constitutive activation of a proto-oncogene or constitutive inhibition of a gene coding for product suppressing cell growth);
- Ignoring or bypassing the pro-apoptotic signals and/or the checkpoint/s of the cell cycle where the pro-apoptotic decisions are normally made (usually, the G1/S checkpoint) (e.g. loss of functional TP53 gene copies);
- Occurrence of molecular or genomic event/s that confer genome instability, increasing the risk for occurrence of additional mutations (e.g. mutations in genes coding for proteins with roles in the maintenance of genome integrity; accelerated telomere attrition, etc.);
- Deregulation of differentiation (differentiation blockage), producing cells with precursor-like phenotype (typically characterised by high proliferation capacity).
The initial event that triggers cancerous transformation may belongs to any of these four types. The others may add up later in any order or virtually simultaneously.
In normal cells, the damage detection and repair machinery is alerted whenever there is a signal for the presence of damage (priority being given to damage in transcribed genomic regions) and every cycle of division is preceded by extensive damage checks. If the cell has sustained too much damage that cannot be managed by repair mechanisms, it would be instructed to enter permanent replicative senescence and/or routed to the programmed cell death pathway. Each of these safety mechanisms is very efficient, but they may occasionally not recognise an instance of damage or simply miss it. It is believed that a statistical number of mutation events (that is, at least between 3 and 6) must occur in the same cell in order to trigger neoplastic transformation [
2. Cancer may sometimes be caused by defined (and, in many cases, eliminable) environmental factors
there's nothing like tobacco: it is the passion of respectable men;
and the man who lives without tobacco is not worthy to live.
Jean-Baptiste Poquelin Molière, Don Juan,
or The Feast with the Statue (c. 1660).
Cancer has always been a mystery disease and the isolation and the characterisation of the leading pathogenetic factors of tumour growth still present a major challenge to biomedical research. The simple fact that cancer may be caused by exposure to certain exogenous agents has been known for quite some time before the actual pathogenetic mechanisms of cancer were discovered. The earliest attempt for a serious study on the link between factors in the environment and the risk of cancer belongs to Percival Pott (1714–1788), a British surgeon (later knighted), who demonstrated that specific types of cancer were associated with specific occupations. Namely, he found that cancer of the scrotum was almost exclusively seen in chimney-sweeps and named exposure to soot as the culprit [reviewed in
It has been known for quite a long time before the nature of the causative agent was identified, that cervical cancer was more common in married women than in unmarried women that practiced celibacy. In 1842, the Italian physician Domenico Rigoni-Stern published his observations on the epidemiology of cervical cancer, stating that cervical cancer was rather common in women with multiple sexual partners (in his studies – prostitutes); and very rare in women living a life of celibacy (as the author bluntly stated, 'nuns, virgins and spinsters'), except for nuns that had chosen the monastic life in later age [Rigoni-Stern, 1842; reviewed in
Other associations between environmental factors and common cancers were not elucidated until the XX century. For some of them, the connection simply could not have been made earlier. For example, having suntanned complexion, especially for women, has never been considered attractive in Europe (as it was usually considered to be a sign of lower class origin) up until the 20-ties of the XX century, when one of the famous French celebrities of the time, Coco Chanel, had an accidental sunburn during an ocean cruise and decided to show off her new tanned looks instead of trying to conceal them. Very soon, deep tan was considered to be a symbol of health and fitness and 'heliotherapy' was proclaimed to be a cure for all diseases. The finding that skin cancers appeared predominantly on sun-exposed areas of skin was first published in the late 40-ties of the XX century [
Tobacco was first brought into Europe in early XVI century, but the link between tobacco smoking and lung cancer was demonstrated unequivocally only 60 years ago, in 1950 [
The origins of many cancers cannot be unequivocally linked to any environmental factor/s, and 'healthy living' is not a guarantee than one would remain cancer-free until their old age; neither is 'unhealthy living' directly associated with development of cancer. It has been accepted that accumulation of unrepaired DNA damage (in the process of ageing, or for other reasons – e.g. high levels of oxidative stress, defects in recognition and repair of damage, etc.) is a major mechanism for triggering carcinogenesis, with or without the presence of carcinogenic factors of the environment.
3. Cancer cells are not that alien to normal cells
and different except for the similarities.
Thomas Sowell, The Vision of the Anointed (1996).
3.1. Cancer cells share some common characteristics with normal cells
Cancer comes in so many types and the properties of cancer cells may be so dissimilar, that making a unified definition of a cancer cell is not easy. The basic properties of cancer cells may be summarised as follows:
- Cells that are capable of division beyond the Hayflick's limit or may divide indefinitely;
- Cells with metastatic potential (capable of invading and colonising new sites that are a long way away from their place of origin);
- Cell with properties characteristic of undifferentiated cells or of cells at earlier stages of differentiation, that are incapable of terminal differentiation, unless under special circumstances.
In cells undergoing cancerous transformation, the increased proliferation capacity is usually acquired first and the capacity for metastasising adds up later.
The first two properties (high proliferative potential and metastatic potential) are not unique to cancer cells. Other types of cells such as embryonic cells and stem cells also have high proliferative potential (sometimes virtually unlimited, e.g. in cultured pluripotent cells). Some non-cancerous cells are naturally capable of colonising new habitats. For example, transplanted haematopoietic stem cells eventually colonise the bone marrow of the recipient, but they are not transferred to the bone marrow during the actual transplantation. Specifically, the haematopoietic cells are transplanted in the myeloablated recipient by means of a simple IV transfusion of a cell suspension. The cells are then transported by the blood flow to the bone marrow, settle there and replenish the haematopoietic cell niche.
The third characteristic listed above, however – signs of incomplete differentiation – is a defining trait of a cancer cell. As the grade of differentiation of a tumour is a very important characteristic, this will be discussed in more detail below.
There have been reports about stabilised stem cell lines (e.g. lines from induced pluripotent stem cells) exhibiting expression and mRNA profiles characteristic of cancer cells [
3.2. Cancer cells exhibit traits typical of undifferentiated cells or cells at lower differentiation grades
Cancer cells may express proteins or other molecules that are usually part of the expression profile of undifferentiated cells or of partially differentiated precursor cells. The differentiation grade of the tumour is one of the basic characteristics assessed in routine histopathology examination. Differentiation grade may vary from low (undifferentiated or poorly differentiated) to high (moderately to well differentiated). The differentiation grade of tumours is directly associated with their proliferative and/or metastatic potential – the lower the differentiation grade, the higher the aggressiveness of the tumour and, in most cases, the poorer is the prognosis for the patient. The survival rates between patients with poorly differentiated and well differentiated tumours may be drastically different, even for the same type of tumour. For example, there are forms of leukemia with minimal differentiation that are very aggressive, and there are leukemias with higher grade of differentiation that may develop slowly or even run a chronic course. This is easily understandable, as the further the cell has gone on the path of differentiation, the lower its proliferative potential usually becomes. Terminally differentiated cells usually have a very limited capacity for division, if at all (for more detail, see 'Cancer stem cells' below). Also, higher grade of differentiation usually means less capacity for invasion of distant locations and infiltration of other tissues (metastasis). Some of the proteins expressed by cancer cells and characteristic of the undifferentiated state are positive regulators of cell cycle (for example, growth factor receptors, receptor-associated kinases, or other signalling molecules); substances degrading basal laminae and/or stimulators of angiogenesis, facilitating the colonisation of distant sites.
Some (but not all) cancer cells may be specifically stimulated towards differentiation. This is usually accompanied with drastic reduction of the proliferation potential of the tumour cells (respectively, the aggressiveness of the tumour). Induced differentiation is sometimes used as a therapeutic approach (differentiation therapy), especially in haematological cancer. Agents known to induce differentiation in cancer cells in vitro as well as in vivo are, for example, trans-retinoic acid in the treatment of leukemia [
Some differentiating agents (e.g. hormones) are usually efficient in certain types of tumours only, as they alone express the relevant receptor. For example, estrogens and androgens are usually used in the treatment of tumours occurring in tissues dependent on the respective hormone – e.g. the mammary gland, the prostate gland, the ovaries and the endometrium. Estrogen was shown to sensitise some types of tumour cells (e.g. breast and cervical cancer cells) to cytotoxic treatments [
Trans-retinoic acid and its derivatives, 5-azacytidine, and other differentiating agents have a wider spectrum of action and may be applicable for induction of growth arrest in more than one type of tumour. For example, trans-retinoic acid is used in the treatment of leukemia, but also in squamous skin cancer. This is likely to be related to the multiple targets for the action of the drug. For example, trans-retinoic acid promotes differentiation of leukemia cells but also suppresses the expression of anti-apoptotic factors of the BCL-2 protein family [
4. Basic mechanisms of cancer
4.1. 'Double-hit' and 'multiple-hit' mechanism of tumorigenesis
The classical model of Knudson (1971) presents tumorigenesis as a process dependent on genetic (deterministic) factors as well as environmental (stochastic) factors. A basic schematic of the double-hit mechanism is presented on Fig. 1.
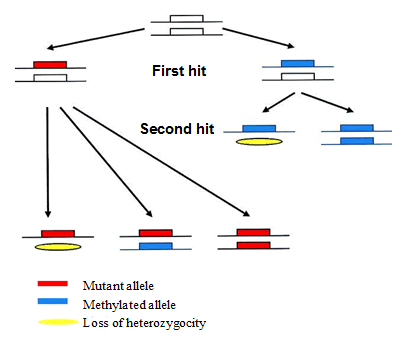
The theoretical concept behind the 'double-hit' mechanism is that no single event may result in cancer. This is valid even when there is a genetic factor conferring cancer-proneness, as Knudson's model was initially developed for a heritable cancer – namely, retinoblastoma. For example, carrying a mutation in the BRCA1 or BRCA2 gene/s may eventually result in breast and/or ovarian cancer, but not before a second mutation occurring on somatic level had knocked out the intact gene copy as well. Indeed, from purely statistical point of view, the risk for somatic inactivation of the second BRCA1 gene copy by random mutagenesis in cells that already have one defective gene copy of the BRCA1 gene is much higher than in cells with both copies intact. The second 'hit', however, may or may not occur, as two 'hits' at the same target is an unlikely event. Even among carriers of BRCA1 mutations, the penetrance of familial breast cancer rarely exceeds 90%, which means that in about 1 in 10 of proven carriers associated cancers do not develop (although part of these 10% may be attributed to lack of accurate data). Later, Knudson's double-hit model was developed further by Vogelstein & Kinzler, who postulated that a statistical number of DNA mutations (generally no less than 3, usually more – 5 or 6) must occur in the same cell in order to transform it to a cancerous cell (multiple-hit model) [
The mechanism of accumulation of multiple 'hits' by random mutagenesis is currently believed to be one of the primary mechanism for triggering cancer growth [
Cancer development may be modified (prevented, slowed down or stimulated) by many factors, some of which are genetic and others are factors of the environment. The two types of factors may be equally important for the development of cancer. For example, it has been shown that homozygotes by a relatively common polymorphism – the 83bp insertion allele in the XPC gene, were at increased risk for development of squamous cell carcinoma and adenocarcinoma of the lung [
The 'random hit' mechanisms of mutagenesis may play a role in other cancer-related processes as well – for example, in development of resistance to anticancer drugs (for more information, see "Bases of cancer resistance to drugs" below.
It has been proposed that genomic instability in some cancers (specifically bone cancer, but possibly some cases of breast cancer as well) does not occur as a result of randomly occurring mutations which happen to 'hit' a crucial gene, but in a single catastrophic event, termed 'chromothripsis', affecting regions on one or several chromosomes 24,25]. According to the authors, the affected region is literally shattered into fragments, some of which (but not all) are subsequently assembled together again by the cellular machinery for DNA repair. Since it is not possible to determine the 'correct' sequence of the genomic fragments, these are patched together in a more or less random order using the only possible way – the mechanism of NHEJ, which is inherently error-prone [
4.2. Should I stay or should I go? Deciding the fate of a new cell
The "50:50" rule
Generally, the daughter cells resulting from the division of a cell are very similar (virtually identical) with regard to their properties, the distribution of the cellular components, and the fate of the individual cells. The latter means that if more than one alternative route is available for a newly formed daughter cell, each of the daughter cells has an equal chance of taking any of these routes. In some types of undifferentiated cells (e.g. stem cells, cancer cells), a specific phenomenon may be observed during cell division. Namely, the daughter cells resulting from division in these types of cells exhibit different properties and may have very dissimilar fates. The two daughter cells often contain different amounts of specific cell compounds and may be intended to take different routes once the division is complete. In stem cells this usually means that one of the two daughter cells (or, more accurately said, 50% of the daughter cells in the population of dividing stem cells in a tissue) were destined from the very start of the cell division to retain the characteristics of the original stem cell; whereas the other 50% were destined to take the route of differentiation. Thus, with every cell division, the stem cell population is replenished, on the one hand, and a differentiating precursor cell is produced, on the other hand. One division of the original stem cell eventually results in production of many specialised cells, as the precursor cell usually undergoes multiple divisions before it eventually enters the replicative arrest that is typical of terminally differentiated cells. The daughter cells that would take on the role of stem cell of the tissue would preserve the characteristic high potential for proliferation and the hyperplastic state of their chromatin; while those that were destined to become differentiated cells would typically lose their capacity for division at some point in the course of differentiation and significant proportion of their chromatin would become condensed and transcriptionally inactive.
Deciding which of the two daughter cells would retain the stemness qualities and which would take the differentiation route is a complex process. It may be directed by exogenous or endogenous factors. Among the former is, for example, the contact with the cell niche. The daughter cells may be positioned differently relative to the cell niche where the mother cell belonged to (one in direct contact with the niche, the other away from it). An important factor of endogenous origin may be, for example, unequal distribution of cell components (mRNA, proteins, membrane-limited compartments, even cell organelles, e.g. mitochondria). DNA may also be distributed in an unequal manner between the daughter cells, as it has been demonstrated that in some cell types the replicated DNA molecules are segregated asymmetrically during division, with all those containing the original 'maternal' DNA strand always dispatched to one of the daughter cells, and those that had been synthesised using a copy made during the previous cycle of division – to the other daughter cell (see below).
Not all cell types employ the 50:50 rule of cell division, but cells in tissues with rapid cell turnover often do. Such are, for example, the adult stem cells in the basal (germinative) layer of mammalian skin. Dead keratinised cells from the upper layers of the skin are regularly sloughed off and must be promptly replaced in order to preserve the skin integrity. Epithelial stem cells generally produce progeny in compliance to the 50:50 rule, except in specific cases that take priority over it. For example, in deep penetrating skin injury the stem cells from the basal layer in the regions adjacent to the injury migrate to the injury site and start dividing, so that eventually new skin grows over the injured site. This means that at some point the progeny of an adult epithelial stem cell must have been comprised predominantly of cells retaining the stemness characteristics, as the epithelial stem cell population at the injury site must be re-established in order to ensure normal skin growth. After the stem cell niche had been replenished, the production of epithelial cells would comply with the 50:50 rule again.
If, for some reason, the cells of the basal layer of the epidermis start cycling faster than usual, this normally would be compensated by acceleration of the process of differentiation of precursor cells. This is exactly the case with some hyperkeratotic states of the skin and mucosa, e.g. skin warts and condylomata acuminata caused by infection with human papillomavirus (usually of the types 6 and 11). In hyperkeratotic epithelia, however, the 50:50 rule is still observed (one cell retaining the stemness qualities, including the proliferation capacity; the other becoming differentiated, gradually losing its ability to divide), only the pace of cell cycle is altered by the virus in order to produce quickly as many viral particles as possible.
Infection with HPV may sometimes cause shifting of division of epithelial cells away from the 50:50 rule towards predominant production of cells with high proliferative potential and hyperplastic chromatin – in other words, cells with phenotype typical of the undifferentiated cells [
Asymmetric segregation of daughter DNA molecules during cell division as an anti-cancer mechanism
DNA is replicated by a semi-conservative mechanism, meaning that the double strands in each of the daughter cells are made of one strand of 'parental' origin (serving as a template in replication) and one newly synthesised strand. This is valid for every chromosome in eukaryotic cells. In most cell types, the physical segregation of the two sets of chromosomes in the end of mitosis is at random, that is, each of the daughter cells may receive a chromosome of each pair which contains the 'original' ('ancestral') strand of DNA or a strand that has been copied from the complementary strand, a product of copying in the previous cell cycle.
The likelihood that a cell would receive a set of chromosomes containing the 'original' DNA strand purely by chance is very low indeed, in the order of one in about ten million. In some types of cells, however, e.g. in rodent embryonic cells in primary tissue culture, the chromosomes carrying the same 'ancestral' DNA strand have been shown to be always selectively targeted together to the same cell [
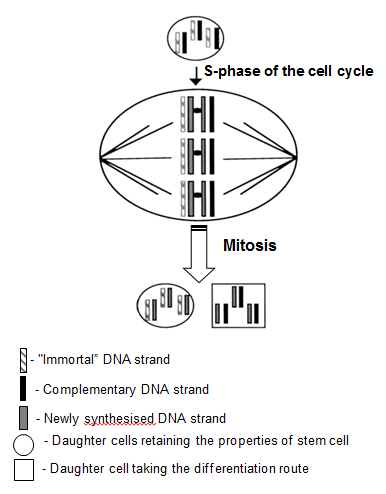
The hypothesis about the "immortal DNA strand" was first formulated in 1975 by the British physician John Cairns [
The mechanisms of DNA replication are inherently error-prone although the error rate is normally very low. If the DNA of a stem cell becomes altered, the alterations would be passed to the cell's progeny and would very likely be multiplied further during subsequent division. This may have various harmful consequences. As the DNA of adult stem cells accumulates errors, their potential for production of new precursor cells gradually declines (one of the hallmarks of ageing); or they may become transformed and start producing mutant cell progeny (for more information, see "Cancer stem cells" below). By asymmetric segregation of the 'original' and the 'copied copy' DNA strands between daughter cells, all the mutations that had previously occurred because of replication errors would presumably be passed onto the cell that is destined to take the differentiation route. Indeed, this 'mutation-laden' cell would divide several (or more) times before it becomes a specialised cell, but the number of divisions would be finite. It is likely that the impending replicative senescence would induce permanent cell cycle arrest before the cell has accumulated enough mutations to become transformed. The daughter cell that retains the stemness qualities would inherit the 'original' blueprint from the mother cell and pass it on in the next division cycle to the next daughter cell destined to remain a stem cell, thus minimising the risk of accumulation of mutations in the stem cell population of the tissue. This makes perfect sense, as most somatic cells in the adult body are replaceable, but the stem cell populations in adult tissues are supposed to last a lifetime.
Not all stem cells employ the mechanism of asymmetric segregation of DNA. For example, the distribution of chromosomes containing the 'immortal' and the 'copied copy' DNA strands during division of haematopoietic stem cells was found to be almost completely random [
5. Cellular genes coding for products that play a role in tumorigenesis
To God I'm the loyal opposition.
Woody Allen, Stardust Memories (1980)
Capacity for unlimited proliferation is one of the main distinguishing traits of cancer cells. Mutations affecting the cellular genes coding for proteins with functions in the regulation of the cell cycle make up for the majority of the somatic 'hits' that eventually trigger tumorigenesis. Depending on the type of regulatory function that these proteins may have in the progression in the cell cycle (positive – stimulating cell proliferation or negative – suppressing cell division), a carcinogenic mutation may produce constitutive and/or ectopic activation of a gene product or, alternatively, suppress the expression and/or reduce or altogether abolish the activity of a gene product. Some pro-carcinogenic mutations do not alter the regulation of the progression through the cell cycle but, rather, increase the overall mutation rate in the genome, creating favourable background for occurrence of other mutations. At present, the most commonly used type of classification of genes and gene products, mutations in which play a role in carcinogenesis, refers the genes and the encoded proteins to one of three major groups. The criterion for differentiation between the three groups is whether they stimulate or suppress cell proliferation (directly or indirectly) or work by destabilisation of genome integrity. The structure of the classification is presented below, and major representatives of each class of pro-carcinogenic genes and proteins are listed.
5.1. Proto-oncogenes
Wild type proto-oncogenes (c-onc) are, in fact, the normal cellular genes coding for products functioning in positive regulation of the cell cycle as signalling or effector molecules. The products of proto-oncogenes may directly stimulate cell proliferation (for example, growth factors) or may be capable of inducing the expression of numerous downstream genes that are directly involved in the stimulation of the cell proliferation and/or the progression through the cell cycle (transcription factors), or may ensure the uneventful passing through cell cycle checkpoints (e.g. components of the cyclin-CDK regulatory system). Proto-oncogenes may also inhibit (directly or indirectly) the expression of genes that are responsible for triggering the pathways of cell cycle arrest, DNA repair and/or apoptosis in damaged cells.
Major human proto-oncogenes are commonly systematised into several sub-groups, depending on their functions (Table 1).
| Function of the protein product of the proto-oncogene |
Examples |
|---|---|
| Growth factors |
c-SIS (platelet-derived growth factor B chain, PDGF2) c-INT2 (fibroblast growth factor 3) c-HST (fibroblast growth factor 4) |
| Receptors with tyrosine kinase activity | Epidermal growth factor receptor (EGFR, ERBB1) Platelet-derived growth factor receptor (PDGFR1) Vascular endothelial growth factor receptors 1 and 2 (VEGFR1 and VEGFR2) c-ROS c-MET c-KIT c-RET |
| Non-receptor tyrosine kinase | c-ABL c-FYN c-LYN c-SRC |
| GTP-binding proteins | c - RAS (K-RAS, H-RAS, N-RAS) |
| G-protein–coupled Receptors | c-MAS |
| Cytoplasmic serine/threonine kinases and their regulatory subunits | c-RAF c-MOS |
| Components of the cyclin-CDK regulatory system | CCND1 (cyclin D1, PRAD1) |
| Nuclear proteins - DNA binding factors, including transcription factors |
c-REL gene family (including NF- Kb) c-MYC c-FOS c-JUN c-MAF |
| Suppressors of apoptosis | BCL-2 gene family members coding for proteins with anti-apoptotic properties (BCL-2, BCL-B, BCL-W, BCL-XL, etc.). |
Most of the cellular proto-oncogenes have viral homologues. A viral proto-oncogene is usually a fragment of a normal cellular proto-oncogene fused with the viral genome, which may become integrated in the genome of the virus-infected cell and/or may be transported around the genome. Virus genomes may integrate into the genome of the host cell, then excise out of the genome and re-integrate elsewhere, sometimes by a 'cut and paste' mechanism and sometimes leaving a copy of the viral genome behind. The excision of a virus genome copy is not always accurate, and recombination may occur between regions of homology belonging to different viral copies integrated in different locations of the genome. Thus, virus genomes may leave fragments of their own DNA sequence in the host cell's DNA and/or fragments of the genome of the host cells may be picked up during virus genome excision, becoming an integral part of the nucleic acid of the virus. The genomic sequences that are carried around by viruses may be functionally active (e.g. complete coding sequences of genes or DNA copies of mRNA transcripts), or may be activated after their translocation – for example, when placed by viral genome integration into an actively transcribed region of the genome of the host cell; in a location within the range of action of strong promoters or enhancers, etc. If the incorporated genomic sequence contains potentially functional fragments of a proto-oncogene, the integration of the virus genome copy into the genome of the host cell may result in upregulation of cell proliferation that may, eventually, produce overt cancer. Insertion of viral genome copies in the host cell's DNA may also cause illegitimate activation of the cellular gene/s in the vicinity of the integration site, because of disruption of the physiological control over their expression. Such modes of pro-carcinogenic action are prerogative of viruses with life cycle that includes integration in the genome of the infected cell (transforming viruses, oncoviruses). Commonly, these are viruses with RNA genomes that propagate via DNA intermediates (retroviruses), although DNA-based viruses such as the papillomavirus and the Epstein-Barr virus may also have transforming properties, as they are capable of integration into the genome of host cells.
Most viral proto-oncogenic homologues were discovered before the eponymous cellular proto-oncogene, as part of the genome of a virus with known oncogenic properties. For example, the src gene, responsible for the transforming function of Rous sarcoma virus (RSV) was differentiated from the gag, pol and env genes of the virus in 1976 [
Pro-carcinogenic mutations in cellular proto-oncogenes are usually of the gain-of-function type, resulting in expression of the gene product at abnormally high levels and/or ectopic and/or constitutive expression (this is also the principle of action of most viral proto-oncogenes). Carriership of inherited gain-of-function mutations in normal cellular proto-oncogenes is usually incompatible with life. The majority of gain-of-function mutations are, therefore, of somatic origin. Exceptions to this are, for example, the heterozygous mutations in the c-RET proto-oncogene. c-RET codes for a receptor protein kinase, involved in the development of the embryonic intestine and kidneys and the enteric nervous system [
Loss-of-function mutations in proto-oncogenes may sometimes be heritable in man. As could be expected, these are usually not associated with cancer-proneness but, rather, with defects in cell migration and distribution of precursor cells in their assigned locations during early embryonic life. For example, carriership of heterozygous mutations in the region encoding the kinase domain of the human c-KIT gene, decreasing the response to KIT ligand binding, may be associated with the phenotype of piebaldism [
Sometimes, same type of mutations in the same proto-oncogene may be associated with different phenotypes. For example, both gain-of-function and loss-of-function mutations in the c-KIT gene may be associated with familial gastrointestinal stromal cancer [
5.2. Tumour-suppressor genes
The products of tumour-suppressor genes play a role in damage-associated signalling and the induction of cell cycle arrest and/or apoptosis in response to DNA damage or other type of damage. Pro-carcinogenic mutations in tumour-suppressor genes are usually of the loss-of-function type and may be heritable. Individuals that have inherited one defective copy of a tumour-suppressor gene are at risk of developing a tumours in case the intact gene copy is lost or inactivated at somatic level (e.g. via the double-hit mechanism). Inherited mutations in tumour-suppressor genes contribute to carcinogenesis in different ways. For example, they may increase the risk for introduction of 'errors' in DNA during cell division (because of inefficient 'checking' mechanism; inefficient DNA repair and/or inefficient damage-associated signalling); or they may cause suppression of the mechanism for the induction of cell cycle arrest and/or apoptosis in the presence of DNA damage. Some inherited mutations in tumour-suppressor genes may indirectly cause stimulation of cell growth, e.g. by increasing the binding of pro-proliferation factors [
5.3. Mutator genes
The genome of undifferentiated cells (including cancer cells) is changeable in real time (hyperplastic). Some traits may be lost, while other may be newly acquired, and genes and whole gene clusters may be rapidly switched on and off regardless of the requirements of the normal developmental programme. The hyperplasticity of the genome of cancer cells is a crucial part of their capacity to adapt to changing environmental conditions (e.g. colonising new sites and invading tissues different from the tissue of origin of the tumour) and in response to anticancer treatments. Important players in the induction and maintenance of genome hyperplasticity are the products encoded by the so-called 'mutator genes'. The term 'mutator genes' denotes a large group of genes coding for proteins with diverse functions. A common feature of mutator genes is that mutations in them result in a net increase in the overall mutation rate in the genome of the transformed cell. Naturally, this result in increased risk for occurrence of mutations in proto-oncogenes and 'second hits' in loci in which there is already one inactive gene copy of a tumour-suppressor or a mutator gene. Thus, mutations in mutator genes are associated with susceptibility for cancer not because of direct interference in the mechanisms of stimulation or suppression of cell proliferation, but, rather, because they create a favourable environment for occurrence of other pro-carcinogenic mutations. Mutations in mutator genes may occur at somatic level or may be inherited, and the associated diseases and conditions may be transmitted in an autosomal dominant as well as in autosomal recessive manner.
The mutant protein products of mutator genes may contribute to destabilisation of the genome via several different mechanisms. One of these mechanisms is, for example, increasing the risk for introduction of errors during DNA replication. Such are, for example, human genes coding for proteins acting in repair of mismatched bases in DNA – MSH1, MSH6, MLH1, etc. [
The genes coding for the RNA component (TERC) or the protein component (TERT) of the telomerase complex are often denoted as mutator genes as well, as mutations in them (somatic as well as inherited) may contribute to cancer development. The carcinogenic mechanisms associated with deregulation of telomere maintenance may significantly vary. Abnormally short telomeres and free reactive chromosome ends may produce chromosome instability – translocations, chromosome fusion and breakage, etc. Inherited mutations in the TERT and TERC genes associated with disordered telomere elongation may produce dyskeratosis congenita; idiopathic aplastic anaemia or idiopathic pulmonary fibrosis; and, in case of deletion of a larger chromosome region, containing the TERT locus or the whole 5p chromosome arm – the cri-du-chat contiguous gene syndrome [
Ability to re-synthesise telomeric DNA is usually associated with increased capacity for cell proliferation, postponing the onset of replicative senescence; or cell immortalisation. Usually, somatic cells have virtually non-existent telomerase activity and adult stem cells have but a limited capacity for telomere elongation. Re-activation or upregulation of telomerase activity in somatic cells is strongly indicative of cancerous transformation [
Some cancer cells are capable of telomere elongation by an alternative mechanism, based on recombination (alternative lengthening of telomeres, ALT, also called alternative telomere lengthening, ATL). In ALT, it is not the chromosome's own telomeric DNA used as a template for copying during replication, but telomeric DNA from another chromosome. Specifically, single-stranded DNA end from one telomere invades double-stranded DNA of another telomere and uses it as a template for copying, eventually producing telomeric DNA with greater length than initial invading telomere end [
It is currently believed that in about 10–15% of all tumours the telomerase activity is lost at some point, beyond which the telomere length is maintained entirely by ALT [
Other types of mutator genes normally regulate the expression of other genes implicated in the control of cell growth. Deregulation of the function of the mutator genes of this type promotes cancerous transformation by stimulation or inhibition of the transcription of their target genes. For example, mutations in the gene coding for protein kinase C alpha may promote uncontrolled cell growth [
The boundaries between the three categories of cancer-associated genes may sometimes become blurred. For example, some of the 'classic' tumour-suppressor genes (prime examples are TP53, ATM, BRCA1 and BRCA2 genes) may also be classed as mutator genes, as they are involved in the maintenance of genome integrity as well. ATM, for example, is usually considered a mutator gene, as the loss of two gene copies produces genome instability, but is sometimes viewed as a tumour suppressor, as its protein product functions in induction of damage-associated cell cycle arrest and apoptosis. Similarly, mutations in the proto-oncogene c-RET may produce cancer, but it is usually via the double-hit mechanism that is considered typical of tumour-suppressor genes.
Currently, there is yet another classification of cancer-associated genes, assigning 'gatekeeper' functions to tumour-suppressor genes and 'caretaker' functions to mutator genes [
Caretaker genes usually play a role in the maintenance of the genome integrity and DNA repair universally, in all tissues. Their inactivation does not promote cancerous growth directly but, rather, by increasing the likelihood of occurrence of mutations in other genes, including caretaker genes. Examples for caretaker genes are BRCA1 and BRCA2, the XP genes coding for proteins acting in NER, the MLH and MSH genes, encoding proteins functioning in mismatch repair, etc. Again, ATM is usually classed together with the caretaker genes.
6. Bases of cancer resistance to genotoxic drugs
Resistance to anticancer drugs may develop due to various reasons. Among the common causes for development of drug resistance may be upregulation of the expression of gene/s coding for product/s that function in the sequestering of the drug or an active metabolite so that they become unavailable or subthreshold; and/or their rapid clearance, and/or their degradation. Some drugs used in anticancer therapy are metabolised by one or more specific enzyme systems. Thus, resistance to the drug may be induced by simple upregulation of the expression of the respective enzyme/s in the tumour. For example, many antitumour drugs are substrates for the cytochrome 1B1 (CYP1B1) enzyme of the cytochrome Р450 family. Among these are agents with direct genotoxic effect (e.g. inhibitors of topoisomerase II, such as mitoxanthrone); microtubule stabilisers (taxanes); antiestrogens (tamoxifen, flutamide), tyrosine kinase inhibitors (imatinib) and others [
Cancer cells may also physically multiply (amplify) the active gene copies coding for the enzyme or a key subunit of an enzyme that degrades or inactivates in any other manner the active compounds of anticancer drugs. Normally, the expression of such proteins is strictly controlled. The gene amplification ensures that the synthesis of the protein encoded by the amplified gene is constantly kept at a high level. Such is the case, for example, with dihydrofolate reductase (DHFR) gene, encoding an enzyme inactivating methotrexate and other cytostatic drugs [
Cells resistant to anticancer compounds may not rely on detoxification of the active substance/s or metabolite/s but, rather, on decreasing their effective concentrations within the cell. This may be implemented via binding of the compound to drug transporter proteins, such as multidrug resistance proteins (MDPs). MDPs are usually transmembrane proteins with high affinity to different chemical agents. They efficiently bind and export a variety of anticancer compounds outside the cell [reviewed in
The genes coding for MDP may also be subjected to copy number amplification and/or upregulation of expression. For example, multidrug resistance in human cancer cell lines to colchicine, vinca alkaloids and anticancer antibiotics such as adriamycin may be due to increased expression of the multidrug resistance gene MDR1 as well as amplification of the MDR1 gene copies [
Platinum-based anticancer regimens (regardless of whether the platinum compound is used as a single agent or combined with other drugs) are used very often in the treatment of solid tumours because of the high response rates, comparable only to anthracycline-based regimens. Resistance of cancer cells to platinum-based drugs is a specific area, as the mechanisms of detoxification of platinum agents are quite different from these of most anticancer drugs. Generally speaking, out of the wide variety of anticancer compounds, it is only platinum agents that are not 'metabolised' or 'biotransformed', due to their unique structure [
Unlike many drugs administered intravenously, platinum compounds do not rapidly become bound by plasma proteins, but, rather, it is the platinum ions that are bound and transported inside the cell. Cisplatin, the first ever platinum-based drug to be used in treatment of human cancer, and other platinum (II) complexes undergo spontaneous hydrolysis of the two chloride ions in aqueous solutions. The platinum-containing cation is bound by plasma proteins such as albumin, transferrin, and gamma globulin within 2–3 hours after IV administration. The platinum-protein complexes are then slowly cleared, predominantly by renal excretion, over the next several days.
Resistance to platinum regimens may be based on decreased concentration of cisplatin cation in the cell due to lower uptake or to intensified export out of the cell. Until recently, it was believed that platinum ions entered living cells mostly by passive diffusion, but later it was shown that active uptake was also possible. For example, some mammalian copper transporter proteins (e.g. SLC32A1) may transport active platinum compounds as well [
Binding and inactivation (also called 'trapping') of platinum compounds in the cytosol before they have reached the nucleus may also be a mechanism for development of resistance. A number of normal intracellular compounds (e.g. reducing agents such as glutathione and thioredoxin) may bind platinum derivatives in a complex that may subsequently be exported from the cell [
The genotoxic action of platinum derivatives is based on formation of adducts in DNA (dG-dG and dG-dA), mainly between nucleotides in the same DNA strand, but also between different strands. Platinum agents may also cause DNA-protein crosslinks, albeit with lower efficiency [
In mouse models in which the BRCA1 gene was destroyed beyond repair by targeted mutagenesis (e.g. by introducing large deletions), resistance to cisplatin derivatives never developed. Indeed, the tumour never disappeared completely, but always grew back, only to regress promptly after treatment with the same agent [
7. Cancer stem cells
The existence of cancer stem cells has been suspected for some years before the first conclusive evidence appeared in 1997, in research on histopathology of acute myeloid leukemia [
According to the cancer stem cell concept, the tumour mass is made of precursor-like rapidly dividing cells that are usually sensitive to DNA damaging agents. The actual source of these cells, however, has stem cell-like properties; its cells divide slowly and, respectively, are only mildly affected by genotoxic agents, if at all. Less than a dozen cancer stem cells (between 4 and 10) may be sufficient to completely restore the bulk of the tumour [
Existence of cancer stem cells have been definitely proven so far for some cancers only, such as haematological malignancies and some CNS tumours such as gliomas [
8. The final checkpoint – cancer as an adaptive evolutionary mechanism
As we already saw, DNA repair/programmed cell death regulatory mechanisms usually manage DNA damage very efficiently, repairing minor damage and eliminating seriously damaged cells. With ageing, however, the capacity for damage repair and self-renewal of cells and tissues declines and the level of unrepaired damage in the cell (which was low until that moment) begins to rise. Sustaining unrepaired damage in cells that are capable of division increases the risk for introduction of mutations that may be inherited by the cell's progeny. If, as a result of introduction of mutations, the cell acquires the ability to bypass the checkpoints in the cell cycle where the integrity of the genome is assessed and the decisions whether to proceed with the cell cycle are made, it may actually evade the general directive that damaged cells must stop dividing and/or die. As a result of continuous proliferation of the cell carrying the altered genotype and the risk of introducing more mutations, cancer may eventually develop. Certainly, this does not occur overnight, but, rather, as a long-term consequence of expanding and multiplying DNA errors that had occurred a long time ago. The risk of cancer usually rises with age, as a consequence of the decreased capacity for repair and tissue renewal and the longer time during which the organism had been exposed to damaging factors.
It is the simple and inevitable truth that everything that was ever alive must eventually die. This normally occurs after a period of gradual but irreversible decline that is commonly called ageing.). This holds true for all living things on Earth, from the simplest prokaryotes to plants, animals and man. Indeed, bacteria are capable of numerous successive divisions, steadily producing (almost) identical daughter cells. Bacterial daughter cells may be slightly different from the mother cell – because of random mutagenesis; or via exchanges of discrete units of genetic information (e.g. via plasmids) with other bacteria. There is also the error-prone mechanism of translesion DNA copying that may allow prokaryotic cells to survive adverse conditions, even though their DNA is seriously damaged, at the cost of introducing genetic mutations. Compared to the limited number of divisions that eukaryotic cells could typically carry out before the onset of replicative senescence, prokaryotes may be considered very long-lived indeed, practically immortal. Nevertheless, it is now known that prokaryotes may also experience gradual restriction of capacity for growth, resembling the process of ageing. The typical phenotype of a malignant cell comprises the capacity for sustaining multiple divisions (practically indefinitely) and to mutate readily, so as to adapt rapidly to changing conditions (in therapy settings – first-line anticancer therapy; second-, third- and so forth lines of therapy; adjuvant therapy (immunomodulating agents, hormones, biological therapies, etc.)). Cancer cells escape death by actually re-inventing the ancient ways of prokaryotic cells of living and reproducing for a very long time without showing symptoms of ageing. If we may return again to our hypothetical example of a population of complex living beings that do not age (are practically immortal) because of preserved capacity for supplying new cells to make up for those that were lost for any reason throughout their lives we already saw that after long enough time the population would dwindle to a limited number of very old, practically immortal individuals. These individuals were initially very alike in their genetic background but have since accumulated such an enormous mutation burden that they became hardly similar to each other. With a lifespan that long, and given the random nature of spontaneous mutagenesis, each of these individuals would eventually possess its own unique genotype. That would preclude sexual reproduction, as it has very strict requirements for the genetic similarity of the mating individuals. The latter means that they would be unable to sustain the population over time, as old individuals would die, albeit very rarely (e.g. because of a very severe injury) and reproduction would be hardly possible. This is, once again, a dead end, a genetic stagnation, similar to whatever may occur if all errors in DNA were repaired at a 0% error rate, though not because of too little, but of too much genetic diversity. One could hypothesise that ageing (of cells, of tissues, and, ultimately, of the organism) is a safety mechanism put in place by Nature during evolution so as to avoid reversion to the ancient mechanisms (nowadays seen in some prokaryotes only) that may sustain the life of the cell in changing environmental conditions at the expense of introduction of unwarranted genetic variability. Even with all the advancements of modern therapy, cancer eventually kills, as cancer cells are capable of rapid invasion and colonisation of all types of cells and tissues in multicellular organisms, and cancer cells are typically not capable of performing the specialised functions of normal cells. Again, one may speculate that this was the Nature's way to ensure that life based on uncontrolled proliferation and unrestricted mutability is not a viable option for living creatures – at least, not beyond the prokaryotic stage of evolution. Thus, cancer may be viewed as a pre-programmed mechanism, a fail-safe that activates when all other options to prevent immortalisation of cells become unavailable for any reason. Ageing/death of old age, and whenever ageing is not an option, cancer may then be viewed as the large, population-scale equivalents of the cell cycle arrest/DNA repair and programmed cell death mechanisms, designed to work synergistically in order to maintain the continuation of life by sacrificing individuals – be it cells, or living beings. On a cell-sized scale, programmed cell death is the only way to extract a damaged cell from its habitat without lasting damage to its neighbours, so that the tissue, the organ and the individual would continue to live. On a larger scale, death (whether of old age or of cancer) is the only way to sustain life on Earth without permitting the slow, hit-and-miss evolutionary process to accelerate abnormally and/or go astray. One could hypothesise that ageing is the normal 'default' process designed to ensure that (almost) every member of a population is allowed a time period in their life cycle in which they would have an (almost) even chance to contribute their own gene variants into the genetic pool of the population, then die, making space for their successors. The latter would, in turn, grow, reproduce, and eventually die, so that many different genetic combinations would be tried and tested in the course of evolution. Only in the rare cases when a cell manages to successfully escape the many checkpoints and mechanisms that order it to switch off the ageing mechanism beyond a certain point in their timeline and eventually die, thus becoming a threat to the homeostasis of the multicellular organism, may come cancer, which would eliminate the dangerous cell clone by killing the organism that created it in the first place. Cancer may then actually be a preprogrammed mechanism, the ultimate fail-safe placed in all cells of multicellular beings in order to eliminate the risk of creating and propagating genotypes that may potentially threaten the existence of the population, the species and life on Earth as a whole [reviewed in
Certainly, it would be rather simplistic to imagine that the establishment of cancer as the final checkpoint (and any checkpoint or mechanism, for that matter) were premeditated events. Rather, it was a naturally occurring phenomenon that was subsequently selected for during evolution, similarly to sexual reproduction or ageing.
9. Could we really 'cure' cancer?
would be quite pointless. Giving up the fight altogether would be humiliating, and, once
again, pointless. The only viable option before us is to study the laws of Nature
thoroughly, so that we could
have them working for us, not against us.
Arkadiy and Boris Strugatskie, One Billion Years
Before The End Of The World (Definitely Maybe), 1974.
Modern biomedical science is waging a real war on cancer, but while winning the individual fights, it is actually losing the battle in the long term. One in six people in modern societies eventually succumbs to cancer, despite the advances in research and all the achievements of the medicine and the pharmaceutical industry. In the light of the theory that cancer is a natural mechanism that prevents complex living beings from living forever because of risk of reaching an evolutionary dead end, this outcome is hardly unexpected. Medicine, however, has all the potential to become capable, in the near future, to slow down the progression of cancer to terminal phases for long enough so that the life expectancy and the quality of life of the individuals affected by cancer could be comparable to the population average. Indeed, the life expectancy of people diagnosed with some types of cancer has dramatically changed in the last decades, and many could live near-normal, fulfilling lives, or at least be as comfortable as possible under the circumstances. It is very rarely the case, however, that the cancer is truly cured (that is, it never relapses throughout the life of the patient, until eventually they die of old age). Such may be the case with cancers of purely somatic origin that were diagnosed and exhaustively treated at very early stage. Most cancers, however, are never completely eradicated, no matter what treatments are undertaken, but are merely arrested in their progression. Anticancer treatments are usually not a long-term cure, as cancer cells eventually manage to become resistant to all currently available treatments.
Why does the united anti-cancer front always fail in the end? We already saw that cell proliferation is tightly controlled at multiple levels to ensure that there is at least one (preferably more than one) option for induction of cell cycle arrest and/or apoptosis at every level, if DNA damage or other types of damage are present. Cancer cells eventually achieve resistance to all currently known chemotherapeutic agents exactly because of the opportunities for interference in the progression through the cell cycle at different stages. Indeed, the capacity for regulation at almost every step of normal cell proliferation means that there are potential 'control overrides' at any stage. A checkpoint may be evaded, a crucial checkpoint controller may be eliminated (e.g. by introducing inactivating mutations or deletion of the gene copies coding for the wild type protein) or made to work in the opposite direction (e.g. cancer-specific isoforms of various genes coding for proteins directly regulating the progression through the cell cycle). In most cases, however, the abnormal cell is identified quite early in the course of its cancerous transformation by any of these self-same mechanisms for checking the status of DNA and genome integrity; and is usually promptly removed by programmed cell death. It is only very rarely indeed that a cell would accumulate enough mutation events so as to fully unleash its tumorigenic potential.
One of the topics that commonly come up in mass media, usually under the headline of 'sensational news', is 'Scientists invented a miracle drug [or other type of treatment] that would put a stop to cancer'. This rather bold (for a lack of better word) announcement may be followed by a short passage made up of mangled sentences compiled from research reports published in the international scientific data banks (typically ones that came up there years ago) and assembled together in a manner that suggests that this is latest-minute news and that the researchers are only a split second away from making a groundbreaking discovery that would eradicate cancer forever. Many real and very useful findings in the field of biology and medicine have suffered this fate (if we would only care to remember the widely publicised idea of stem cells being a cure for all diseases), and many patients with cancer and their families have learned the hard way that there is no universal remedy for all diseases, except, maybe faith, hope and love. Indeed, it is hardly conceivable from scientific point of view that one could possibly invent 'a cure for leukemia' or 'a cure for breast cancer'. The different types of cancer affecting the same tissue and/or organ may be very different, to the point that the only unifying feature between them may be that they happen to occur in the same location in the body. The misconception that one cure may work for all varieties of the same type of tumour probably stems from the early days of oncology, when the only tool available to the physician for examination of a living patient was observation, and when tumours were classed according to the organ or bodily part that they affect. Often, different forms of the same type of tumour are very dissimilar to one another in respect of their aggressiveness, expression profile, eligibility for treatment with different agents, response to various therapies, and other important characteristics. For example, among leukemias there are very aggressive forms (e.g. acute blast leukemias) which follow a rapidly progressive course despite the modern treatment modalities; and there are indolent forms (e.g. some types of chronic lymphocytic leukemia) that may need treatment only at late stages, if at all. The outcomes of treatment for different varieties of the same generic type of tumour may also greatly vary. For example, the survival rate of Hodgkin's lymphoma, when diagnosed and treated at an early stage, may be between 85 and 98%, while for other types of lymphoma (e.g. angioimmunoblastic T-cell lymphoma) the 5-year survival rate is still around 30% [
Every type of cancer ought to be viewed as a separate entity, with its unique origin, properties and course, and every cancer sufferer must be treated with regard to their own unique nature. A universal 'cure for cancer' as such does not exist at the moment and is unlikely to be ever invented.
Acknowledgеments
This research was supported by Grant No. DFNI-B01/2 at the National Science Fund, Ministry of Education and Science of Republic of Bulgaria.
References
- Bédard A, Parent A. Evidence of newly generated neurons in the human olfactory bulb. Brain Res Dev Brain Res. 2004;151(1-2):159–68.
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–41.
- Kipling MD, Waldron HA. Percivall Pott and cancer scroti. Br J Ind Med. 1975;32(3):244–6.
- Griffiths M. "Nuns, virgins, and spinsters". Rigoni-Stern and cervical cancer revisited. Br J Obstet Gynaecol. 1991;98(8):797–802.
- Versluys JJ. Cancer and occupation in the Netherlands. Br J Cancer. 1949;3(2):161–85.
- Cannon AB. Skin Tumors. Bull N Y Acad Med. 1947;23(3):163–72.
- Wynder EL, Graham, E A. Tobacco smoking as a possible etiologic factor in bronchiogenic carcinoma; a study of 684 proved cases. J Am Med Assoc. 1950;143(4):329–36.
- Gunaratne PH. Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr Stem Cell Res Ther. 2009;4(3):168-77.
- Zhong X, Li N, Liang S, Huang Q, Coukos G, Zhang L. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem. 2010;285(53):41961-71.
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270(2):488-98.
- Arabadjiev B, Petkova R, Chakarov S, Momchilova A, Pankov R. Do we need more human embryonic stem cell lines? Biotechnol Biotechnol Eq. 2010; 24(3):1921-7.
- Masetti R, Vendemini F, Zama D, Biagi C, Gasperini P, Pession A. All-trans retinoic acid in the treatment of pediatric acute promyelocytic leukemia. Expert Rev Anticancer Ther. 2012;12(9):1191–204.
- Fassina G, Aluigi MG, Gentleman S, Wong P, Cai T, Albini A, et al. The cAMP analog 8-Cl-cAMP inhibits growth and induces differentiation and apoptosis in retinoblastoma cells. Int J Cancer. 1997;72(6):1088–94.
- Okumura T. Mechanisms by which thiazolidinediones induce anti-cancer effects in cancers in digestive organs. J Gastroenterol. 2010;45(11):1097–102.
- Tallman MS. Differentiating therapy in acute myeloid leukemia. Leukemia. 1996;10(8):1262–8.
- He Q, Liang CH, Lippard SJ. Steroid hormones induce HMG1 overexpression and sensitize breast cancer cells to cisplatin and carboplatin. Proc Natl Acad Sci U S A. 2000;97(11):5768–72.
- Chau KY, Lam HY, Lee KL. Estrogen treatment induces elevated expression of HMG1 in MCF-7 cells. Exp Cell Res. 1998;241(1):269–72.
- Lovejoy KS, Lippard SJ. Non-traditional platinum compounds for improved accumulation, oral bioavailability, and tumor targeting. Dalton Trans. 2009;(48):10651–9.
- Edvardsson K, Ström A, Jonsson P, Gustafsson J-Å, Williams C. Estrogen receptor β induces antiinflammatory and antitumorigenic networks in colon cancer cells. Mol Endocrinol. 2011;25(6):969–79.
- Andreeff M, Jiang S, Zhang X, Konopleva M, Estrov Z, Snell VE, et al. Expression of Bcl-2-related genes in normal and AML progenitors: changes induced by chemotherapy and retinoic acid. Leukemia. 1999;13(11):1881–92.
- Venitt S. Mechanisms of spontaneous human cancers. Environ Health Perspect. 1996;104 Suppl 633–7.
- López-Cima MF, González-Arriaga P, García-Castro L, Pascual T, Marrón MG, Puente XS, et al. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162.
- Qiu L, Wang Z, Shi X, Wang Z. Associations between XPC polymorphisms and risk of cancers: A meta-analysis. Eur J Cancer. 2008;44(15):2241–53.
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. Elsevier; 2011;144(1):27–40.
- Rajapakse I, Scalzo D, Groudine M. Losing control: cancer's catastrophic transition. Nucleus. 2(4):249–52.
- Chen J-M, Férec C, Cooper DN. Transient hypermutability, chromothripsis and replication-based mechanisms in the generation of concurrent clustered mutations. Mutat Res. 750(1):52–9.
- Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18(11):1630–8.
- Funk JO. Cancer cell cycle control. Anticancer Res. 1999;19(6A):4772–80.
- Petkova R, Tsekov I, Yemendzhiev H, Kalvatchev Z. Let Sleeping Dogs Lie – Unleashing the Transforming Power of Dormant Hpv. Biotechnol Biotechnol Equip. 2012;26(1):2689–94.
- Petkova R, Chelenkova P, Yemendjiev H, Tsekov I, Chakarov St, Kalvachev Zl. HPV has left the building – the absence of detectable HPV DNA and the presence of R allele/s for the P72P polymorphism in the TP53 gene may call for more aggressive therapeutic approach in HPV-associated tumours. Biotechnol Biotechnol Eq. 2013;27(6): 4217-21.
- Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243(4893):934–7.
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505.
- Sun Z-Q, Wang H-J, Zhao Z-L, Wang Q-S, Fan C-W, Fang F. Significance of HPV infection and genic mutation of APC and K-ras in patients with rectal cancer. Asian Pac J Cancer Prev. 2013;14(1):121–6.
- Bissada E, Abboud O, Abou Chacra Z, Guertin L, Weng X, Nguyen-Tan PF, et al. Prevalence of K-RAS Codons 12 and 13 Mutations in Locally Advanced Head and Neck Squamous Cell Carcinoma and Impact on Clinical Outcomes. Int J Otolaryngol. 2013;2013:848021.
- Zehbe I, Rätsch A, Alunni-Fabbroni M, Burzlaff A, Bakos E, Dürst M, et al. Overriding of cyclin-dependent kinase inhibitors by high and low risk human papillomavirus types: evidence for an in vivo role in cervical lesions. Oncogene. 1999;18(13):2201–11.
- Tae Kim Y, Kyoung Choi E, Hoon Cho N, Hung Ko J, Ick Yang W, Wook Kim J, et al. Expression of cyclin E and p27(KIP1) in cervical carcinoma. Cancer Lett. 2000;153(1-2):41–50.
- Li B, Hu Y, Ye F, Li Y, Lv W, Xie X. Reduced miR-34a expression in normal cervical tissues and cervical lesions with high-risk human papillomavirus infection. Int J Gynecol Cancer. 2010;20(4):597–604.
- Mellone M, Rinaldi C, Massimi I, Petroni M, Veschi V, Talora C, et al. Human papilloma virus-dependent HMGA1 expression is a relevant step in cervical carcinogenesis. Neoplasia. 2008;10(8):773–81.
- Anderson S, Shera K, Ihle J, Billman L, Goff B, Greer B, et al. Telomerase activation in cervical cancer. Am J Pathol. 1997;151(1):25–31.
- Resar LMS. The high mobility group A1 gene: transforming inflammatory signals into cancer? Cancer Res. 2010;70(2):436–9.
- Lark KG, Consigli RA, Minocha HC. Segregation of sister chromatids in mammalian cells. Science. 1966;154(3753):1202–5.
- Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115(Pt 11):2381–8.
- Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8(7):677–87.
- Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5(5):e102.
- Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132(4):681–7.
- Karpowicz P, Morshead C, Kam A, Jervis E, Ramunas J, Ramuns J, et al. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170(5):721–32.
- Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics. Cancer Res. 2002;62(23):6791–5.
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255(5505):197–200.
- Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449(7159):238–42.
- Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129(7):1244–7.
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207.
- Parlakian A, Gomaa I, Solly S, Arandel L, Mahale A, Born G, et al. Skeletal muscle phenotypically converts and selectively inhibits metastatic cells in mice. PLoS One. 2010;5(2):e9299.
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 59(4):225–49.
- Duesberg PH, Wang LH, Beemon K, Kawai S, Hanafusa H. Sequences and functions of Rous sarcoma virus RNA. Hamatol Bluttransfus. 1976;19:327–40.
- Beemon K, Hunter T. In vitro translation yields a possible Rous sarcoma virus src gene product. Proc Natl Acad Sci U S A. 1977;74(8):3302–6.
- Le Beau MM, Westbrook CA, Diaz MO, Rowley JD. Evidence for two distinct c-src loci on human chromosomes 1 and 20. Nature. 1984;312(5989):70–1.
- Heisterkamp N, Groffen J, Stephenson JR, Spurr NK, Goodfellow PN, Solomon E, et al. Chromosomal localization of human cellular homologues of two viral oncogenes. Nature. 1982;299(5885):747–9.
- Cockburn JG, Richardson DS, Gujral TS, Mulligan LM. RET-mediated cell adhesion and migration require multiple integrin subunits. J Clin Endocrinol Metab. 2010;95(11):E342–6.
- Eng C, Mulligan LM. Mutations of the RET proto-oncogene in the multiple endocrine neoplasia type 2 syndromes, related sporadic tumours, and hirschsprung disease. Hum Mutat. 1997;9(2):97–109.
- Huang SC, Koch CA, Vortmeyer AO, Pack SD, Lichtenauer UD, Mannan P, et al. Duplication of the mutant RET allele in trisomy 10 or loss of the wild-type allele in multiple endocrine neoplasia type 2-associated pheochromocytomas. Cancer Res. 2000;60(22):6223–6.
- Isozaki K, Terris B, Belghiti J, Schiffmann S, Hirota S, Vanderwinden JM. Germline-activating mutation in the kinase domain of KIT gene in familial gastrointestinal stromal tumors. Am J Pathol. 2000;157(5):1581–5.
- Rothschild G, Sottas CM, Kissel H, Agosti V, Manova K, Hardy MP, et al. A role for kit receptor signaling in Leydig cell steroidogenesis. Biol Reprod. 2003;69(3):925–32.
- Giebel LB, Spritz RA. Mutation of the KIT (mast/stem cell growth factor receptor) protooncogene in human piebaldism. Proc Natl Acad Sci U S A. 1991;88(19):8696–9.
- Spritz RA, Giebel LB, Holmes SA. Dominant negative and loss of function mutations of the c-kit (mast/stem cell growth factor receptor) proto-oncogene in human piebaldism. Am J Hum Genet. 1992;50(2):261–9.
- Pasini B, Borrello MG, Greco A, Bongarzone I, Luo Y, Mondellini P, et al. Loss of function effect of RET mutations causing Hirschsprung disease. Nat Genet. 1995;10(1):35–40.
- Nishida T, Hirota S, Taniguchi M, Hashimoto K, Isozaki K, Nakamura H, et al. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet. 1998;19(4):323–4.
- Penchovsky R, Chacarov S, Genova G. The tumour suppressor mutation 1(2)gd in drosophila melanogaster elevates the binding activity of the transcription factors AP-1. Compt Rend Bulg Acad Sci. 1998; 51(7-8):83-6.
- Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, et al. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428(6980):328-32.
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75(5):1027–38.
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368(6468):258–61.
- Ball SE, Gibson FM, Rizzo S, Tooze JA, Marsh JC, Gordon-Smith EC. Progressive telomere shortening in aplastic anemia. Blood. 1998;91(10):3582–92.
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413(6854):432–5.
- Armanios M, Chen J-L, Chang Y-PC, Brodsky RA, Hawkins A, Griffin CA, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci U S A. 2005;102(44):15960–4.
- Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci U S A. 2007;104(18):7552–7.
- Ohyashiki JH, Ohyashiki K, Iwama H, Hayashi S, Toyama K, Shay JW. Clinical implications of telomerase activity levels in acute leukemia. Clin Cancer Res. 1997;3(4):619–25.
- Engelhardt M, Ozkaynak MF, Drullinsky P, Sandoval C, Tugal O, Jayabose S, et al. Telomerase activity and telomere length in pediatric patients with malignancies undergoing chemotherapy. Leukemia. 1998;12(1):13–24.
- Nowak T, Januszkiewicz D, Zawada M, Pernak M, Lewandowski K, Rembowska J, et al. Amplification of hTERT and hTERC genes in leukemic cells with high expression and activity of telomerase. Oncol Rep. 2006;16(2):301–5.
- Heselmeyer-Haddad K, Sommerfeld K, White NM, Chaudhri N, Morrison LE, Palanisamy N, et al. Genomic amplification of the human telomerase gene (TERC) in pap smears predicts the development of cervical cancer. Am J Pathol. 2005;166(4):1229–38.
- Nagel I, Szczepanowski M, Martín-Subero JI, Harder L, Akasaka T, Ammerpohl O, et al. Deregulation of the telomerase reverse transcriptase (TERT) gene by chromosomal translocations in B-cell malignancies. Blood. 2010;116(8):1317–20.
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–14.
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21(4):598–610.
- Wu G, Jiang X, Lee W-H, Chen P-L. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63(10):2589–95.
- Jiang W-Q, Zhong Z-H, Henson JD, Neumann AA, Chang AC-M, Reddel RR. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol Cell Biol. 2005;25(7):2708–21.
- Zhong Z-H, Jiang W-Q, Cesare AJ, Neumann AA, Wadhwa R, Reddel RR. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282(40):29314–22.
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73(2):347–60.
- Royle NJ, Méndez-Bermúdez A, Gravani A, Novo C, Foxon J, Williams J, et al. The role of recombination in telomere length maintenance. Biochem Soc Trans. 2009;37(Pt 3):589–95.
- Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11(5):319–30.
- Durant ST. Telomerase-independent paths to immortality in predictable cancer subtypes. J Cancer. 2012;3:67–82.
- Praskova M, Kalenderova S, Miteva L, Poumay Y, Mitev V. Dual role of protein kinase C on mitogen-activated protein kinase activation and human keratinocyte proliferation. Exp Dermatol. 2002;11(4):344–8.
- Yamasaki T, Takahashi A, Pan J, Yamaguchi N, Yokoyama KK. Phosphorylation of Activation Transcription Factor-2 at Serine 121 by Protein Kinase C Controls c-Jun-mediated Activation of Transcription. J Biol Chem. 2009;284(13):8567–81.
- Ruvolo PP, Deng X, Carr BK, May WS. A functional role for mitochondrial protein kinase Calpha in Bcl2 phosphorylation and suppression of apoptosis. J Biol Chem. 1998;273(39):25436–42.
- Olsen J V, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–48.
- Alvaro V, Lévy L, Dubray C, Roche A, Peillon F, Quérat B, et al. Invasive human pituitary tumors express a point-mutated alpha-protein kinase-C. J Clin Endocrinol Metab. 1993;77(5):1125–9.
- Prévostel C, Martin A, Alvaro V, Jaffiol C, Joubert D. Protein kinase C alpha and tumorigenesis of the endocrine gland. Horm Res. 1997;47(4-6):140–4.
- Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386(6627):761, 763.
- Rochat B. Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism. Clin Pharmacokinet. 2005;44(4):349–66.
- McFadyen MC, Cruickshank ME, Miller ID, McLeod HL, Melvin WT, Haites NE, et al. Cytochrome P450 CYP1B1 over-expression in primary and metastatic ovarian cancer. Br J Cancer. 2001;85(2):242–6.
- Gibson P, Gill JH, Khan PA, Seargent JM, Martin SW, Batman PA, et al. Cytochrome P450 1B1 (CYP1B1) is overexpressed in human colon adenocarcinomas relative to normal colon: implications for drug development. Mol Cancer Ther. 2003;2(6):527–34.
- Bertino J, Göker E, Gorlick R, Li W, Banerjee D. Resistance Mechanisms to Methotrexate in Tumors. Oncologist. 1996;1(4):223–6.
- Pérez-Tomás R. Multidrug resistance: retrospect and prospects in anti-cancer drug treatment. Curr Med Chem. 2006;13(16):1859–76.
- Shen DW, Fojo A, Chin JE, Roninson IB, Richert N, Pastan I, et al. Human multidrug-resistant cell lines: increased mdr1 expression can precede gene amplification. Science. 1986;232(4750):643–5.
- Yabuki N, Sakata K, Yamasaki T, Terashima H, Mio T, Miyazaki Y, et al. Gene amplification and expression in lung cancer cells with acquired paclitaxel resistance. Cancer Genet Cytogenet. 2007;173(1):1–9.
- Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4(4):307–20.
- Safaei R, Howell SB. Copper transporters regulate the cellular pharmacology and sensitivity to Pt drugs. Crit Rev Oncol Hematol. 2005;53(1):13–23.
- Hall MD, Okabe M, Shen D-W, Liang X-J, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535.
- Kruh GD. Lustrous insights into cisplatin accumulation: copper transporters. Clin Cancer Res. 2003;9(16 Pt 1):5807–9.
- Komatsu M, Sumizawa T, Mutoh M, Chen ZS, Terada K, Furukawa T, et al. Copper-transporting P-type adenosine triphosphatase (ATP7B) is associated with cisplatin resistance. Cancer Res. 2000;60(5):1312–6.
- Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22(47):7369–75.
- Zaludová R, Zákovská A, Kasparková J, Balcarová Z, Kleinwächter V, Vrána O, et al. DNA interactions of bifunctional dinuclear platinum(II) antitumor agents. Eur J Biochem. 1997;246(2):508–17.
- Borst P, Rottenberg S, Jonkers J. How do real tumors become resistant to cisplatin? Cell Cycle. 2008;7(10):1353–9.
- Dhillon KK, Swisher EM, Taniguchi T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011;102(4):663–9.
- Rottenberg S, Borst P. Drug resistance in the mouse cancer clinic. Drug Resist Updat. 15(1-2):81–9.
- Ikeda H, Matsushita M, Waisfisz Q, Kinoshita A, Oostra AB, Nieuwint AWM, et al. Genetic reversion in an acute myelogenous leukemia cell line from a Fanconi anemia patient with biallelic mutations in BRCA2. Cancer Res. 2003;63(10):2688–94.
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–7.
- Schoppy DW, Ruzankina Y, Brown EJ. Removing all obstacles: a critical role for p53 in promoting tissue renewal. Cell Cycle. 2010;9(7):1313–9.
- Masters JR, Foley CL, Bisson I, Ahmed A. Cancer stem cells. BJU Int. 2003;92(7):661–2.
- Passegué E, Jamieson CHM, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci U S A. 2003;100 Suppl 11842–9.
- Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15(9):1010–2.
- Wang JCY, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15(9):494–501.
- Barami K. Relationship of neural stem cells with their vascular niche: implications in the malignant progression of gliomas. J Clin Neurosci. 2008;15(11):1193–7.
- Venere M, Fine HA, Dirks PB, Rich JN. Cancer stem cells in gliomas: identifying and understanding the apex cell in cancer's hierarchy. Glia. 2011;59(8):1148–54.
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–10.
- Tirino V, Camerlingo R, Franco R, Malanga D, La Rocca A, Viglietto G, et al. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur J Cardiothorac Surg. 2009;36(3):446–53.
- Chakarov S, Petkova R. The final checkpoint - cancer as an adaptive evolutionary mechanism. Biotechnol Biotechnol Eq. 2014;29(1).
- Federico M, Rudiger T, Bellei M, Nathwani BN, Luminari S, Coiffier B, et al. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: analysis of the international peripheral T-cell lymphoma project. J Clin Oncol. 2013;31(2):240–6.
- Wilson WH, Dunleavy K, Pittaluga S, Hegde U, Grant N, Steinberg SM, et al. Phase II study of dose-adjusted EPOCH and rituximab in untreated diffuse large B-cell lymphoma with analysis of germinal center and post-germinal center biomarkers. J Clin Oncol. 2008;26(16):2717–24.